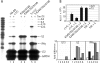Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase - PubMed (original) (raw)
Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase
Arnab Nayak et al. J Biol Chem. 2009.
Abstract
The family of NFAT (nuclear factor of activated T-cells) transcription factors plays an important role in cytokine gene regulation. In peripheral T-cells NFATc1 and -c2 are predominantly expressed. Because of different promoter and poly(A) site usage as well as alternative splicing events, NFATc1 is synthesized in multiple isoforms. The highly inducible NFATc1/A contains a relatively short C terminus, whereas the longer, constitutively expressed isoform NFATc1/C spans an extra C-terminal peptide of 246 amino acids. Interestingly, this NFATc1/C-specific terminus can be highly sumoylated. Upon sumoylation, NFATc1/C, but not the unsumoylated NFATc1/A, translocates to promyelocytic leukemia nuclear bodies. This leads to interaction with histone deacetylases followed by deacetylation of histones, which in turn induces transcriptionally inactive chromatin. As a consequence, expression of the NFATc1 target gene interleukin-2 is suppressed. These findings demonstrate that the modification by SUMO (small ubiquitin-like modifier) converts NFATc1 from an activator to a site-specific transcriptional repressor, revealing a novel regulatory mechanism for NFATc1 function.
Figures
FIGURE 1.
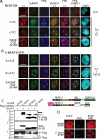
NFATc1/C harbors three SUMO consensus motifs which facilitate interaction with Ubc9. A, schematic representation of NFATc1 isoforms and the SUMO consensus motifs (Ψ-Lys-_X_-Glu; Ψ= isoleucine/leucine/valine) shown by arrows. The short isoforms harbor only the common Lys-349 site, whereas NFATc1/C contains sites at Lys-702 and -914 in addition. Lysines to arginine exchanges create ΔSUMO mutations. The mutation of both C-terminal lysines is designated as K702R/K914R and of all three sites as K349R/K702R/K914R. An indication for the B-term and CC-term peptides are given. TAD, transactivation domain; RSD, Rel similarity domain; NLS, nuclear localization signal; NES, nuclear export signal; SRR and SP, Ser/Thr phosphorylation sites, respectively. B, NFATc1/C physically interacts with Ubc9 in vivo. HA-tagged c1/C-coding vector was transfected into 293T HEK cells, either in combination with pcDNA expressing FLAG-tagged Ubc9 or FLAG only. After 6 h of stimulation with T/I and a total of 48 h, protein lysates were prepared and subjected to anti-FLAG IP followed by immunoblot detection with anti-HA. Whole cell lysates (WCL) were also directly analyzed by immunoblotting with anti-HA and anti-FLAG, respectively. C, the SUMO site-deficient mutant K349R/K702R/K914R cannot recruit Ubc9. Procedures were the same as in B, except that K349R/K702R/K914R was included.D, the SUMO site at Lys-702 is the most relevant. Procedures were the same as in C but including all different SUMO site deficient mutants.E, the C terminus of NFATc1/C is sufficient for binding Ubc9. Procedures were the same as before, but EYFP-tagged NFATc1/C or only the C terminus were analyzed. F, both the NFATc1/B specific (B-term) and the NFATc1/C only (CC-term) C termini interact with Ubc9. Procedures were as before, but by use of constructs with nuclear localization signal-estrogen receptor fusions (estrogen receptor α), which are released for nuclear localization by addition of 4-hydroxytamoxifen.*, heavy or light chain of the precipitation antibody.
FIGURE 2.
NFATc1/C is sumoylated in vivo. A, NFATc1/C was sumoylated. HA-tagged c1/C-coding vector was transfected into 293T HEK cells either with FLAG-tagged SUMO1- or FLAG-expressing pcDNA, and cells were treated with T/I for 6 h. Anti-FLAG immunoprecipitation was followed by IB with anti-HA (arrows, SUMO-modified NFATc1/C) and whole cell lysates (WCL) were analyzed for expression by anti-HA and anti-FLAG.B, the SUMO site-deficient mutant K349R/K702R/K914R cannot be sumoylated. Procedures were as in A, but K349R/K702R/K914R was included, and cells were left unstimulated or treated for 6 h as indicated.C, SUMO site mutants reveal site-specific sumoylation pattern of c1/C. HA-tagged c1/C and different SUMO site mutants of c1/C were subjected to IP as in A, and expression was compared by anti-HA (NFAT) and anti-FLAG (SUMO) IB in whole cell lysates. D, NFATc1 precipitation reveals sumoylated c1/C. Procedures were as in A, except NFATc1/C-ER-stimulated by Tm + T/I for 4 h, and either anti-ER or anti-NFATc1 was used for IP and anti-SUMO1 for IB. E, in CD4+ T-cells endogenous NFATc1 is sumoylated. Nuclei from CD4+ T-cells after 7 days restimulated or not by ionomycin or T/I for 5 h were subjected to IP and IB as in D; IP-anti-NFATc1 + IB-anti-SUMO1 (arrow, sumoylated NFATc1/C, fourth lane, no antibody for IP); reprobe of IB by anti-NFATc1, IB of nuclear lysates (NL) by anti-SUMO1 and anti-NFATc1.
FIGURE 3.
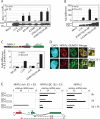
Sumoylation directs NFATc1 into PML-nbs. A, the sumoylatable long isoform colocalizes with PML-nbs. Human A3.01 cells retrovirally infected with constructs expressing c1/A-, c1/C-, and K349R/K702R/K914R-ER and selected by zeocin were left untreated or stimulated (w/o) with Tm+T/I for 4 h. IF was performed with anti-ERα (to detect exogenous NFATc1), anti-SUMO1, and anti-PML followed by laser scanning confocal microscopy. B, the fusion with SUMO directs any NFATc1 isoform to PML-nbs. 293T HEK cells were transfected with SUMO(S)-c1/A, -c1/C, and -K349R/K702R/K914R fused to EYFP at the C terminus and treated with T/I+CaCl2 for 1 h. IF with anti-SUMO1 and anti-PML was performed for triple localization analyzed by confocal microscopy. The scale bar represents 10 μm. C, the C-terminal peptide is sumoylated. 293T HEK cells were transfected with FLAG-SUMO1 and C terminus-specific peptides fused to ER with the addition of an nuclear localization signal. Analyzes were performed as for Fig. 1_F_. WCL, whole cell lysates. D, the sumoylated C terminus is not sufficient for recruitment to PML-nbs. Procedures were as in A but after infection of A3.01 with a retroviral vector expressing the C terminus-specific peptide fused to EYFP. DAPI, 4′,6-diamidino-2-phenylindole;IRES, internal ribosome entry site.
FIGURE 4.
Sumoylation represses NFATc1 transcriptional activity. A, IL-2 promoter activity is increased upon non-sumoylation of NFATc1/C. 293T HEK cells were transfected with 10 μg of pHA-NFATc1/C-EGZ or ΔSUMO mutants along with 1 μg of a luciferase reporter plasmid driven by the IL-2 promoter. After 36 h luciferase activity was measured from cells that were either left untreated or treated with T/I for 16 h. Data are represented as the mean ± S.E. To check for equal NFAT expression, Western blots were performed from these protein extracts. RLU, relative light units. B, fusion with SUMO1 impairs NFATc1 activity. Procedures are as in A, except SUMO1(S)-fused NFATc1 (-ER) clones were transfected in addition. Indicated is Fold difference in relation to basal activity (mock = 1). C, sumoylated NFATc1/C suppresses secretion of endogenous IL-2. Retrovirally infected A3.01 cells were stimulated by Tm+T/I for 48 h, and supernatants were analyzed by enzyme-linked immunosorbent assay. D, nTreg cells exhibit a predominant colocalization of NFATc1 with SUMO. CD4+ and CD4+CD25+ T-cells were isolated from mouse lymph nodes and stimulated with plate-bound anti-CD3 and anti-CD28 antibodies for 3 days. After further 2 days of resting cells were restimulated with T/I for 6 h. IF was performed with anti-NFATc1 and anti-SUMO1 followed by confocal microscopy. The scale bar represents 5 μm. DAPI, 4′,6-diamidino-2-phenylindole. E, nTregs mainly express the long isoform NFATc1/C (E2 + E3), initiated at the second promoter, P2, after restimulation. Real-time reverse transcription-PCR was performed from freshly isolated (0 h), stimulated as in D (5d), and restimulated with plate-bound anti-CD3 and anti-CD28 antibodies for 3 h (5d + 3 h) (no bars= not detectable). After normalization to the expression of HGPRT mRNA, relative expression levels of NFATc1 E1+E3, NFATc1 E2+E3, and NFATc2 mRNAs were calculated, whereas the value gained from unstimulated CD4+ (NFATc1 E2+E3 or NFATc2) was taken as 1.
FIGURE 5.
Sumoylation of NFATc1 can both suppress and enhance lymphokine induction. A, IL-2 mRNA is repressed, but RNA levels of IL-13 and IFNγ are further up-regulated upon sumoylation of NFATc1. 5 μg RNA of retrovirally infected EL-4 cells (NFATc1/C-ER, its ΔSUMO and SUMO fusion mutants) were subjected to RNase protection assay after stimulating the cells with Tm+T/I for 24 h using the mCK1 template set. Also a graph of phosphor-imaging values is given of three independent experiments where 1-6 correspond to the lanes of the autoradiography. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, the presence of SUMO suppresses the transactivation potential of NFATc1. As in Fig. 4_A_, except murine EL-4 cells were transfected and plasmids expressing FLAG (F) or FLAG-SUMO1 (S) were cotransfected with those encoding NFATc1/C, K349R/K702R/K914R, or empty vector (HA) along with an IL-2-luciferase plasmid.
FIGURE 6.
Sumoylation promotes NFATc1 association with class I and II HDACs, which mediates histone deacetylation at the IL-2 promoter. A, HDAC2 interacts with NFATc1/C but not K349R/K702R/K914R. 293T HEK cells transfected with constructs coding for Myc-tagged HDAC 2 and different forms of HA-tagged NFATc1 were treated with T/I for 6 h. Anti-Myc IP was followed by anti-HA antibody for IB, and the expression levels were analyzed directly in whole cell lysates by anti-HA and anti-Myc. In lane 4 specificity was checked by the use of full-length c-Myc-ER construct in place of Myc-tagged HDAC2. WCL, whole cell lysates. B, also, HDAC1 and 4 interact with NFATc1/C. Procedures were as in A, except HDAC1 and HDAC4 constructs were included. C, SUMO fusion recruits HDAC2 to NFATc1. Procedures were as in A, but NFATc1-ER constructs were transfected, and IB was performed by anti-ER. D, sumoylatable NFATc1/C deacetylates histone H3 at the IL-2 promoter. ChIP assay of retrovirally infected A3.01 cells (Figs. 3 and 4), stimulated with Tm+T/I for 6 h, was performed with anti-Ac-H3. DNA from precipitated chromatin was PCR-amplified by using human IL-2 or actin promoter-specific primers. This is one of three independent experiments, and the result was also confirmed in murine EL-4 cells. E, NFATc1 isoforms bind equally to the IL-2 promoter. Procedures were as in C, except anti-ERα was (subsequently) applied to the same cell lysates. F, NFATc1/C expression mediates HDAC2 binding to the IL-2 promoter. Procedures were as in_C_, except cells were only stimulated for 3 h, and anti-HDA2 was applied.
FIGURE 7.
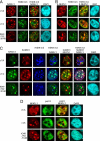
Sumoylation directs NFATc1 into transcriptionally inactive chromatin. A, 293T HEK cells were transfected with plasmids encoding pHA-NFATc1/A, -c1/C, and -K349R/K702R/K914R and stimulated with T/I+CaCl2 for 1 h. IF was performed to detect NFATc1 with anti-HA and heterochromatin with anti-H3K9m3. Colocalization was analyzed by confocal microscopy. The scale bar represents 10 μm. B, 293T HEK cells were transfected with plasmids encoding SUMO1(S)-c1/A, -c1/C, and - K349R/K702R/K914R, which were C-terminally fused to EYFP. Cells were stimulated with T/I+CaCl2 for 1 h. IF was performed with anti-H3K9m3, and colocalization of SUMO-fused NFATc1 (S-NFATc1) and heterochromatin were analyzed by confocal microscopy. C, FLAG-SUMO1 was transfected along with c1/A, c1/C, and K349R/K702R/K914R plasmids, C-terminally fused to EYFP into 293T HEK cells, and stimulated with T/I+CaCl2 for 1 h followed by IF with anti-SUMO and anti-H3K9m3. Tri-colocalization of NFATc1, SUMO1, and H3K9m3 was analyzed by confocal microscopy. D, without sumoylation, NFATc1 colocalizes to hotspots of transcription. Transfection and stimulation was as in C. To reveal the colocalization pattern of NFATc1 and pol II-bodies, IF was performed with anti-pol II antibodies. The scale bar represents 10 μm.DAPI, 4′,6-diamidino-2-phenylindole.
Similar articles
- Sumoylation of a small isoform of NFATc1 is promoted by PIAS proteins and inhibits transactivation activity.
Kim ET, Kwon KM, Lee MK, Park J, Ahn JH. Kim ET, et al. Biochem Biophys Res Commun. 2019 May 21;513(1):172-178. doi: 10.1016/j.bbrc.2019.03.171. Epub 2019 Apr 2. Biochem Biophys Res Commun. 2019. PMID: 30952432 - Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation.
Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, Davidson I, Dejean A. Neyret-Kahn H, et al. Genome Res. 2013 Oct;23(10):1563-79. doi: 10.1101/gr.154872.113. Epub 2013 Jul 26. Genome Res. 2013. PMID: 23893515 Free PMC article. - Inhibiting ubiquitination causes an accumulation of SUMOylated newly synthesized nuclear proteins at PML bodies.
Sha Z, Blyszcz T, González-Prieto R, Vertegaal ACO, Goldberg AL. Sha Z, et al. J Biol Chem. 2019 Oct 18;294(42):15218-15234. doi: 10.1074/jbc.RA119.009147. Epub 2019 Jul 8. J Biol Chem. 2019. PMID: 31285264 Free PMC article. - Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization.
Shih HM, Chang CC, Kuo HY, Lin DY. Shih HM, et al. Biochem Soc Trans. 2007 Dec;35(Pt 6):1397-400. doi: 10.1042/BST0351397. Biochem Soc Trans. 2007. PMID: 18031230 Review. - The Regulation of Chromatin by Dynamic SUMO Modifications.
Wilson NR, Hochstrasser M. Wilson NR, et al. Methods Mol Biol. 2016;1475:23-38. doi: 10.1007/978-1-4939-6358-4_2. Methods Mol Biol. 2016. PMID: 27631795 Review.
Cited by
- Regulatory T-Cell-Mediated Suppression of Conventional T-Cells and Dendritic Cells by Different cAMP Intracellular Pathways.
Rueda CM, Jackson CM, Chougnet CA. Rueda CM, et al. Front Immunol. 2016 Jun 2;7:216. doi: 10.3389/fimmu.2016.00216. eCollection 2016. Front Immunol. 2016. PMID: 27313580 Free PMC article. Review. - NFAT proteins: emerging roles in cancer progression.
Mancini M, Toker A. Mancini M, et al. Nat Rev Cancer. 2009 Nov;9(11):810-20. doi: 10.1038/nrc2735. Nat Rev Cancer. 2009. PMID: 19851316 Free PMC article. Review. - The E3 SUMO ligase PIASγ is a novel interaction partner regulating the activity of diabetes associated hepatocyte nuclear factor-1α.
Kaci A, Keindl M, Solheim MH, Njølstad PR, Bjørkhaug L, Aukrust I. Kaci A, et al. Sci Rep. 2018 Aug 24;8(1):12780. doi: 10.1038/s41598-018-29448-w. Sci Rep. 2018. PMID: 30143652 Free PMC article. - Regulatory T cells facilitate the nuclear accumulation of inducible cAMP early repressor (ICER) and suppress nuclear factor of activated T cell c1 (NFATc1).
Vaeth M, Gogishvili T, Bopp T, Klein M, Berberich-Siebelt F, Gattenloehner S, Avots A, Sparwasser T, Grebe N, Schmitt E, Hünig T, Serfling E, Bodor J. Vaeth M, et al. Proc Natl Acad Sci U S A. 2011 Feb 8;108(6):2480-5. doi: 10.1073/pnas.1009463108. Epub 2011 Jan 24. Proc Natl Acad Sci U S A. 2011. PMID: 21262800 Free PMC article. - The transcriptional coactivator Bob1 promotes the development of follicular T helper cells via Bcl6.
Stauss D, Brunner C, Berberich-Siebelt F, Höpken UE, Lipp M, Müller G. Stauss D, et al. EMBO J. 2016 Apr 15;35(8):881-98. doi: 10.15252/embj.201591459. Epub 2016 Mar 8. EMBO J. 2016. PMID: 26957522 Free PMC article.
References
- Weaver, C. T., Harrington, L. E., Mangan, P. R., Gavrieli, M., and Murphy, K. M. (2006) Immunity 24 677-688 - PubMed
- Serfling, E., Berberich-Siebelt, F., Chuvpilo, S., Jankevics, E., Klein-Hessling, S., Twardzik, T., and Avots, A. (2000) Biochim. Biophys. Acta 1498 1-18 - PubMed
- Hogan, P. G., Chen, L., Nardone, J., and Rao, A. (2003) Genes Dev. 17 2205-2232 - PubMed
- Macian, F. (2005) Nat. Rev. Immunol. 5 472-484 - PubMed
- Ranger, A. M., Hodge, M. R., Gravallese, E. M., Oukka, M., Davidson, L., Alt, F. W., de la Brousse, F. C., Hoey, T., Grusby, M., and Glimcher, L. H. (1998) Immunity 8 125-134 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous