Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties - PubMed (original) (raw)
Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties
Piotr Neumann et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Borna disease virus (BDV) is a neurotropic enveloped RNA virus that causes a noncytolytic, persistent infection of the central nervous system in mammals. BDV belongs to the order Mononegavirales, which also includes the negative-strand RNA viruses (NSVs) Ebola, Marburg, vesicular stomatitis, rabies, mumps, and measles. BDV-M, the matrix protein (M-protein) of BDV, is the smallest M-protein (16.2 kDa) among the NSVs. M-proteins play a critical role in virus assembly and budding, mediating the interaction between the viral capsid, envelope, and glycoprotein spikes, and are as such responsible for the structural stability and individual form of virus particles. Here, we report the 3D structure of BDV-M, a full-length M-protein structure from a nonsegmented RNA NSV. The BDV-M monomer exhibits structural similarity to the N-terminal domain of the Ebola M-protein (VP40), while the surface charge of the tetramer provides clues to the membrane association of BDV-M. Additional electron density in the crystal reveals the presence of bound nucleic acid, interpreted as cytidine-5'-monophosphate. The heterologously expressed BDV-M copurifies with and protects ssRNA oligonucleotides of a median length of 16 nt taken up from the expression host. The results presented here show that BDV-M would be able to bind RNA and lipid membranes simultaneously, expanding the repertoire of M-protein functionalities.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
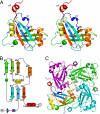
Overall structure of BDV-M. (A) The BDV-M monomer, rainbow-colored from the N to the C terminus. The position of the bound ribonucleotide (cytidine–5′-monophosphate) is indicated in stick representation. (B) Topology of the BDV-M monomer with residue numbers of secondary structure elements (β-strands as arrows, α-helices as cylinders) colored according to A. (C) Ribbon diagram of the (crystallographic) BDV-M tetramer, highlighting the tesselation of the L-shaped monomers.
Fig. 2.
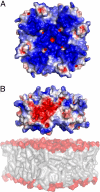
Surface properties of the putative membrane-binding face of the BDV-M tetramer. (A) The electrostatic surface potential reveals this face to be highly basic (areas colored in white, red, and blue denote neutral, negative, and positive potential contoured at 0, − 3, and +3 kT/e, respectively). View is rotated 180° around the horizontal axis relative to Fig. 1_C_. (B) Side view of the tetramer, obtained by rotating (A) by 90° about a horizontal axis, depicting BDV-M approaching a phospholipid membrane (Lower). The alternating surface charges (acidic/basic) of the tetramer edges would facilitate lateral assembly to form planar arrays.
Fig. 3.
The nucleotide binding site in BDV-M. (A) Experimental electron density (omit map contoured at 3 σ) with refined monoribonucleotide cytidine-5′-monophosphate at the interface between 2 monomers (view and colors as in Fig. 1_C_). The pyrimidine ring is sandwiched between the side chains of Phe 37-and His-112 and is held in position through hydrogen bonds to the main chain of Phe-37 and the carboxamide group of the neighboring Asn-115 (distances in Å). In addition, the O2′ atom of a riboxynucleotide sugar moiety would hydrogen-bond to the side chain of Gln 36. (B) Electrostatic potential surface of BDV-M (contour levels as in Fig. 2) with refined RNA trinucleotide (CCC). Note the alternative positions of the phosphate groups in the crystallographically 4-fold averaged structure.
Fig. 4.
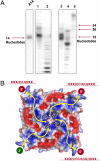
Heterologously-expressed BDV-M binds ssRNA oligonucleotides. (A) Urea-PAGE of isolated nucleic acid from purified BDV-M tetramer. Left lane, defined 14-nt polyadenine marker; lane 1, isolated RNA labeled at the 5′ terminus using PNK; lane 2, as lane 1 after RNase T1 digestion; lane 3, isolated RNA labeled at the 3′-terminus using PAP; lane 4, as lane 3 after RNase T1 digestion; lane 5, polyadenine marker. These results prove the copurification of heterologously expressed BDV-M tetramer with RNA oligonucleotides of ≈16 nt. (B) Schematic depiction of possible binding mode of ssRNA. A distinctive basic patch along the tetramer diagonals could accommodate the polyphosphate backbone (yellow arrows), such that an incoming chain (3′-end bottom left) ends with the observed bound nucleotide (center). Assuming a specificity for cytidine/uridine, there are 3 possible exit routes for the 5′-end: (i) at the top left, with 1 pyrimidine base bound near the tetramer axis; (ii) at the top right, with 2 nt; and (iii) at the bottom right, with 3 central bases contributing to specificity. It is not possible to distinguish between them because of the 4-fold crystallographic symmetry.
Similar articles
- Crystal structure of the borna disease virus nucleoprotein.
Rudolph MG, Kraus I, Dickmanns A, Eickmann M, Garten W, Ficner R. Rudolph MG, et al. Structure. 2003 Oct;11(10):1219-26. doi: 10.1016/j.str.2003.08.011. Structure. 2003. PMID: 14527390 - The glycosylated matrix protein of Borna disease virus is a tetrameric membrane-bound viral component essential for infection.
Stoyloff R, Strecker A, Bode L, Franke P, Ludwig H, Hucho F. Stoyloff R, et al. Eur J Biochem. 1997 May 15;246(1):252-7. doi: 10.1111/j.1432-1033.1997.t01-2-00252.x. Eur J Biochem. 1997. PMID: 9210491 - Crystallization and preliminary X-ray analysis of the matrix protein of Borna disease virus.
Kraus I, Scheffczik H, Eickmann M, Kiermayer S, Stubbs MT, Garten W. Kraus I, et al. Acta Crystallogr D Biol Crystallogr. 2002 Aug;58(Pt 8):1371-3. doi: 10.1107/s0907444902010132. Epub 2002 Jul 20. Acta Crystallogr D Biol Crystallogr. 2002. PMID: 12136159 - Nucleocytoplasmic shuttling of viral proteins in borna disease virus infection.
Honda T, Tomonaga K. Honda T, et al. Viruses. 2013 Aug 8;5(8):1978-90. doi: 10.3390/v5081978. Viruses. 2013. PMID: 23965528 Free PMC article. Review. - The atypical strategies used for gene expression of Borna disease virus, a nonsegmented, negative-strand RNA virus.
Schneemann A, Schneider PA, Lipkin WI. Schneemann A, et al. Uirusu. 1995 Dec;45(2):165-74. doi: 10.2222/jsv.45.165. Uirusu. 1995. PMID: 8820535 Review.
Cited by
- Suppression of Borna Disease Virus Replication during Its Persistent Infection Using the CRISPR/Cas13b System.
Sasaki S, Ogawa H, Katoh H, Honda T. Sasaki S, et al. Int J Mol Sci. 2024 Mar 20;25(6):3523. doi: 10.3390/ijms25063523. Int J Mol Sci. 2024. PMID: 38542493 Free PMC article. - Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Tarbouriech N, Chenavier F, Kawasaki J, Bachiri K, Bourhis JM, Legrand P, Freslon LL, Laurent EMN, Suberbielle E, Ruigrok RWH, Tomonaga K, Gonzalez-Dunia D, Horie M, Coyaud E, Crépin T. Tarbouriech N, et al. Viruses. 2022 Oct 26;14(11):2358. doi: 10.3390/v14112358. Viruses. 2022. PMID: 36366462 Free PMC article. - Dissecting the Subcellular Localization, Intracellular Trafficking, Interactions, Membrane Association, and Topology of Citrus Leprosis Virus C Proteins.
Leastro MO, Kitajima EW, Silva MS, Resende RO, Freitas-Astúa J. Leastro MO, et al. Front Plant Sci. 2018 Sep 11;9:1299. doi: 10.3389/fpls.2018.01299. eCollection 2018. Front Plant Sci. 2018. PMID: 30254655 Free PMC article. - Human Parainfluenza Virus Type 3 Matrix Protein Reduces Viral RNA Synthesis of HPIV3 by Regulating Inclusion Body Formation.
Zhang S, Cheng Q, Luo C, Qin Y, Chen M. Zhang S, et al. Viruses. 2018 Mar 11;10(3):125. doi: 10.3390/v10030125. Viruses. 2018. PMID: 29534486 Free PMC article. - Molecular Requirements for Self-Interaction of the Respiratory Syncytial Virus Matrix Protein in Living Mammalian Cells.
Trevisan M, Di Antonio V, Radeghieri A, Palù G, Ghildyal R, Alvisi G. Trevisan M, et al. Viruses. 2018 Mar 3;10(3):109. doi: 10.3390/v10030109. Viruses. 2018. PMID: 29510513 Free PMC article.
References
- de la Torre JC. Reverse-genetic approaches to the study of Borna disease virus. Nat Rev Microbiol. 2006;4:777–783. - PubMed
- Richt JA, Rott R. Borna disease virus: A mystery as an emerging zoonotic pathogen. Vet J. 2001;161:24–40. - PubMed
- Staeheli P, Sauder C, Hausmann J, Ehrensperger F, Schwemmle M. Epidemiology of Borna disease virus. J Gen Virol. 2000;81:2123–2135. - PubMed
- Dürrwald R, Kolodziejek J, Herzog S, Nowotny N. Meta-analysis of putative human bornavirus sequences fails to provide evidence implicating Borna disease virus in mental illness. Rev Med Virol. 2007;17:181–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials