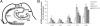The unfolded protein response is activated in pretangle neurons in Alzheimer's disease hippocampus - PubMed (original) (raw)
The unfolded protein response is activated in pretangle neurons in Alzheimer's disease hippocampus
Jeroen J M Hoozemans et al. Am J Pathol. 2009 Apr.
Abstract
Accumulation of misfolded proteins in the endoplasmic reticulum triggers a cellular stress response called the unfolded protein response (UPR) that protects the cell against the toxic buildup of misfolded proteins. Previously, we reported that UPR activation is increased in Alzheimer's disease (AD) patients. How the UPR relates to the pathological hallmarks of AD is still elusive. In the present study, the involvement of UPR activation in neurofibrillary degeneration in AD was investigated. Immunoreactivity for the phosphorylated UPR activation markers pancreatic ER kinase (pPERK), eukaryotic initiation factor 2alpha, and inositol-requiring enzyme 1alpha was observed in hippocampal neurons associated with granulovacuolar degeneration. The percentage of pPERK-immunoreactive neurons was increased in AD cases compared with nondemented control cases and with the Braak stage for neurofibrillary changes. Although absent from neurofibrillary tangles, pPERK immunoreactivity was most abundant in neurons with diffuse localization of phosphorylated tau protein. Additional analyses showed that pPERK immunoreactivity was associated with ubiquitin and the ubiquitin binding protein p62. A strong co-occurrence of immunoreactivity for both pPERK and glycogen synthase kinase 3beta in neurons was also observed. Together, these data indicate that UPR activation in AD neurons occurs at an early stage of neurofibrillary degeneration and suggest that the prolonged activation of the UPR is involved in both tau phosphorylation and neurodegeneration in AD pathogenesis.
Figures
Figure 1
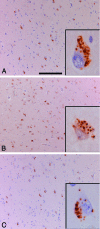
Immunohistochemical localization of pPERK, peIF2α, and pIRE1α in AD hippocampus. A: pPERK is detected by immunohistochemistry in pyramidal neurons in the hippocampus. In this case a part of the subiculum of an AD case is shown. The inset shows the cellular localization of pPERK, which is present in granules that can be defined as GVD. B: peIF2α immunohistochemistry on the same AD case and area as shown in A. peIF2α can also be detected in GVD granules (inset). C: pIRE1α can also be detected in pyramidal neurons in the hippocampus of AD cases (shown is subiculum). Similar granular structures were detected as observed with pPERK and peIF2α immunohistochemistry (inset). Scale bars: 200 μm (A–C); 20 μm (insets).
Figure 2
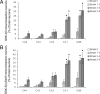
Quantitative analyses of pPERK immunoreactivity. A: Neurons immunoreactive for pPERK were quantified in different regions of the hippocampus. Arrows indicate the borders between different hippocampal regions. B: The percentages of neurons immunoreactive for pPERK were quantified in nondemented control cases, AD cases, AD cases with Lewy body variant (AD/LBV), and dementia cases with a vascular insufficiency (DEM/VASC). Data are expressed as mean ± SEM. *P < 0.05 compared with control levels in same region.
Figure 3
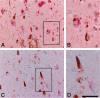
Co-localization of pPERK with markers of the ubiquitin proteasome system. A: Double immunohistochemistry for pPERK (red) and ubiquitin (brown). B: Magnification of inset indicated in A. No co-localization is present between pPERK with ubiquitin inclusions. There is an increased association of pPERK-positive neurons with intra- and extracellular ubiquitin immunoreactive granules. C: Double immunohistochemistry for pPERK (red) and p62 (brown). There is no co-localization between pPERK- and p62-immunoreactive inclusions. D: Magnification of inset as indicated in C. Scale bars: 100 μm (A, C); 50 μm (B, D).
Figure 4
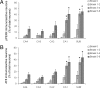
pPERK and AT8 immunoreactivity in different stages of pathology. A: pPERK immunoreactivity in different regions of the hippocampus increases with the Braak score for NFT pathology. B: Similar results are observed with AT8 immunoreactivity. AT8-immunoreactive neurons included immunoreactive NFT as well as diffuse appearance of AT8 immunoreactivity in neurons. Data are expressed as mean ± SEM. *P < 0.05 compared with Braak stage 0 in the same region.
Figure 5
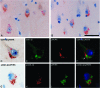
Double immunohistochemistry and spectral imaging analyses for pPERK and phosphorylated tau protein. A: Double immunolabeling for pPERK (brown) and AT8 (red). B and C: Immunohistochemical signals were spectrally unmixed and shown as red for AT8 and green for pPERK. D: Merge of these signals indicates no spatial co-localization. Arrow indicates the absence of pPERK in tangle-bearing neuron. E: Double immunohistochemistry for pPERK (brown) and AT100 (red). F and G: Immunohistochemical signals for AT8 (red) and pPERK (green) were spectrally unmixed. H: Merge of signals for pPERK and AT100 indicate no co-occurrence of pPERK and AT100 in tangle-bearing neuron (arrow). I: Double immunolabeling for pPERK (brown) and AT270 (red). J and K: Immunohistochemical signals for AT270 (red) and pPERK (green) were spectrally unmixed. L: Merge of signal for AT270 and pPERK. Arrow indicates the low amount of pPERK in tangle-bearing neuron. Sections were counterstained with hematoxylin indicated as blue color in merged pictures.
Figure 6
Double immunohistochemistry and spectral imaging analyses for pPERK, GSK-3β, and pGSK-3β. A: Double immunohistochemistry for pPERK (red) and GSK-3β (brown). B: Double immunohistochemistry for pPERK (red) and pGSK-3β (brown). Immunohistochemical signals for pPERK (red) and GSK-3β (brown) (C) were spectrally unmixed in green for GSK-3β (D) and red for pPERK (E). F: Colors were merged with the addition of spectrally unmixed hematoxylin staining indicated in blue. Arrow indicates that GSK-3β in these granules does not co-localize with pPERK. Immunohistochemical signals for pPERK (red) and pGSK-3β (brown) (G) were spectrally unmixed in red for pGSK-3β (H) and green for pPERK (I). J: Colors were merged with the addition of spectrally unmixed hematoxylin staining indicated in blue. Scale bars: 50 μm (A, B); 30 μm (C–J).
Figure 7
GSK-3β and GSK-3β pSer9 immunoreactivity in different stages of pathology. GSK-3β immunoreactivity (A) and GSK-3β pSer9 immunoreactivity (B) in different regions of the hippocampus increases with the Braak score for NFT pathology. Data are expressed as mean ± SEM. *P < 0.05 compared with Braak stage 0 in the same region.
Similar articles
- Activation of the unfolded protein response and granulovacuolar degeneration are not common features of human prion pathology.
Wiersma VI, van Hecke W, Scheper W, van Osch MA, Hermsen WJ, Rozemuller AJ, Hoozemans JJ. Wiersma VI, et al. Acta Neuropathol Commun. 2016 Oct 28;4(1):113. doi: 10.1186/s40478-016-0383-7. Acta Neuropathol Commun. 2016. PMID: 27793194 Free PMC article. - The unfolded protein response is activated in the olfactory system in Alzheimer's disease.
Murray HC, Dieriks BV, Swanson MEV, Anekal PV, Turner C, Faull RLM, Belluscio L, Koretsky A, Curtis MA. Murray HC, et al. Acta Neuropathol Commun. 2020 Jul 14;8(1):109. doi: 10.1186/s40478-020-00986-7. Acta Neuropathol Commun. 2020. PMID: 32665027 Free PMC article. - The unfolded protein response is associated with early tau pathology in the hippocampus of tauopathies.
Nijholt DA, van Haastert ES, Rozemuller AJ, Scheper W, Hoozemans JJ. Nijholt DA, et al. J Pathol. 2012 Apr;226(5):693-702. doi: 10.1002/path.3969. Epub 2012 Feb 17. J Pathol. 2012. PMID: 22102449 - Current advances on different kinases involved in tau phosphorylation, and implications in Alzheimer's disease and tauopathies.
Ferrer I, Gomez-Isla T, Puig B, Freixes M, Ribé E, Dalfó E, Avila J. Ferrer I, et al. Curr Alzheimer Res. 2005 Jan;2(1):3-18. doi: 10.2174/1567205052772713. Curr Alzheimer Res. 2005. PMID: 15977985 Review. - Activation of the unfolded protein response is an early event in Alzheimer's and Parkinson's disease.
Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Scheper W. Hoozemans JJ, et al. Neurodegener Dis. 2012;10(1-4):212-5. doi: 10.1159/000334536. Epub 2012 Feb 1. Neurodegener Dis. 2012. PMID: 22302012 Review.
Cited by
- The unfolded protein response in Alzheimer's disease.
Cornejo VH, Hetz C. Cornejo VH, et al. Semin Immunopathol. 2013 May;35(3):277-92. doi: 10.1007/s00281-013-0373-9. Epub 2013 Apr 23. Semin Immunopathol. 2013. PMID: 23609500 Review. - IRE1α-XBP1 Affects the Mitochondrial Function of Aβ25-35-Treated SH-SY5Y Cells by Regulating Mitochondria-Associated Endoplasmic Reticulum Membranes.
Chu B, Li M, Cao X, Li R, Jin S, Yang H, Xu L, Wang P, Bi J. Chu B, et al. Front Cell Neurosci. 2021 Mar 25;15:614556. doi: 10.3389/fncel.2021.614556. eCollection 2021. Front Cell Neurosci. 2021. PMID: 33841100 Free PMC article. - The Preventive Effects of Salubrinal against Pyrethroid-Induced Disruption of Adult Hippocampal Neurogenesis in Mice.
Toltin AC, Belkadi A, Gamba LM, Hossain MM. Toltin AC, et al. Int J Mol Sci. 2023 Oct 26;24(21):15614. doi: 10.3390/ijms242115614. Int J Mol Sci. 2023. PMID: 37958604 Free PMC article. - Ginkgolide B‑induced AMPK pathway activation protects astrocytes by regulating endoplasmic reticulum stress, oxidative stress and energy metabolism induced by Aβ1‑42.
Wang J, Ding Y, Zhuang L, Wang Z, Xiao W, Zhu J. Wang J, et al. Mol Med Rep. 2021 Jun;23(6):457. doi: 10.3892/mmr.2021.12096. Epub 2021 Apr 21. Mol Med Rep. 2021. PMID: 33880582 Free PMC article. - UBXN1 maintains ER proteostasis and represses UPR activation by modulating translation.
Ahlstedt BA, Ganji R, Mukkavalli S, Paulo JA, Gygi SP, Raman M. Ahlstedt BA, et al. EMBO Rep. 2024 Feb;25(2):672-703. doi: 10.1038/s44319-023-00027-z. Epub 2024 Jan 2. EMBO Rep. 2024. PMID: 38177917 Free PMC article.
References
- Forman MS, Lee VM, Trojanowski JQ. ‘Unfolding’ pathways in neurodegenerative disease. Trends Neurosci. 2003;26:407–410. - PubMed
- Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. - PubMed
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. - PubMed
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. - PubMed
- Hoozemans JJ, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, Eikelenboom P, Scheper W. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol (Berl) 2005;110:165–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous