JNets: exploring networks by integrating annotation - PubMed (original) (raw)
JNets: exploring networks by integrating annotation
Jamie I Macpherson et al. BMC Bioinformatics. 2009.
Abstract
Background: A common method for presenting and studying biological interaction networks is visualization. Software tools can enhance our ability to explore network visualizations and improve our understanding of biological systems, particularly when these tools offer analysis capabilities. However, most published network visualizations are static representations that do not support user interaction.
Results: JNets was designed as a network visualization tool that incorporates annotation to explore the underlying features of interaction networks. The software is available as an application and a configurable applet that can provide a flexible and dynamic online interface to many types of network data. As a case study, we use JNets to investigate approved drug targets present within the HIV-1 Human protein interaction network. Our software highlights the intricate influence that HIV-1 has on the host immune response.
Conclusion: JNets is a software tool that allows interaction networks to be visualized and studied remotely, from within a standard web page. Therefore, using this free software, network data can be presented in an enhanced, interactive format. More information about JNets is available at http://www.manchester.ac.uk/bioinformatics/jnets.
Figures
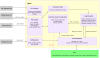
Figure 1
Diagrammatic representation of the the JNets system. This diagram describes the conceptual flow of information through the software.
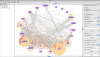
Figure 2
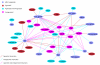
The JNets user interface. The main interface and network visualization panel from the JNets application is shown. JNets is displaying the HIV-1 human PPI network, from HHPID [6,7]. On the left is the legend panel showing the node and edge groups. The menu bar at the top is customizable. Standard drop-down menus can be enabled or disabled and new menus with user-defined network views can be added, defined in the configuration file. On the right the subgroup edit and analysis panel is shown, through which the user can tailor the visualization, explore subgroup annotation and add further annotation to the network.
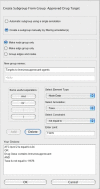
Figure 3
The JNets subgroup creation interface. This interface is used to create a new subgroups from already existing ones. The upper half of this panel is used to select some simple options about the subgroups being created, such as whether the grouping should be made automatically or manually, what the name of new subgroups should be and whether new a new node group, edge group, or both should be created. The lower half of this panel is used to select the annotation and set the filters through which new subgroups will be created.
Figure 4
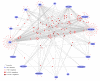
The HIV-1-host, drug-target interaction network. This is the whole network that was used for all analyses in our case study. Using the annotation that accompanies nodes and edges, JNets can filter this network to create more focussed visualizations, for example, the networks in figures 5-7.
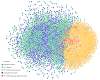
Figure 5
HIV-1 interacting drug target genes. This network shows 237 human genes that encode products that are HIV-1 interacting and FDA-approved drug targets. HIV-1 elements are labelled. Human genes are colored according to the number of distinct HIV-1 elements with which they share an interaction (darker = more). The layout of this network was achieved by manually positioning the HIV-1 nodes, locking them in position and allowing the JNets spring layout to reposition the remaining human gene nodes. As a result, human gene nodes with multiple interactors are drawn to the centre of the network. From this visualization it is clear that many HIDTs interact with multiple HIV-1 elements and that certain HIV-1 elements are responsible for many more HIV-HIDT interactions than others. Such observations can be investigated in greater detail using JNets. For example, 114 from 237 of these human genes interact with more than one HIV-1 element. This is significantly more than would be expected at random (p < 0.001).
Figure 6
Drug-target network showing all immunosuppressant target HIV-1 interacting genes. The drug nodes lie in an arc around the top of the network. Between these two groups are the human gene nodes, colored according to the action HIV-1 has upon them. Four types of action are defined: agonised [7], antagonised [11], agonised and antagonised [12] and neutral/unspecified [5]. These distinctions are derived from the interaction description supplied with each HHPID HIV-1-host interaction. HIV-1 elements are shown at the bottom and are labelled. The human genes shown in this network are likely to perform a significant role in the immune system to be targeted by immunosuppressive drugs. From this visualization it is clear that HIV-1 targets many of the same proteins as immunosuppressant drugs.
Figure 7
HIV-1 host network showing immunosuppressive agent target genes. Human genes that are both HIV-1 interacting and targeted by immunosuppressive agents are shown. Only those genes that are explicitly agonised or antagonised by HIV-1 have been included; human genes are colored according to this action. Seven host genes are agonised, eleven are antagonised and twelve are both agonised and antagonised. These distinctions are derived from the interaction description supplied with each HHPID HIV-1-host interaction.
Similar articles
- BisoGenet: a new tool for gene network building, visualization and analysis.
Martin A, Ochagavia ME, Rabasa LC, Miranda J, Fernandez-de-Cossio J, Bringas R. Martin A, et al. BMC Bioinformatics. 2010 Feb 17;11:91. doi: 10.1186/1471-2105-11-91. BMC Bioinformatics. 2010. PMID: 20163717 Free PMC article. - VisANT: an online visualization and analysis tool for biological interaction data.
Hu Z, Mellor J, Wu J, DeLisi C. Hu Z, et al. BMC Bioinformatics. 2004 Feb 19;5:17. doi: 10.1186/1471-2105-5-17. BMC Bioinformatics. 2004. PMID: 15028117 Free PMC article. - Integrated web visualizations for protein-protein interaction databases.
Jeanquartier F, Jean-Quartier C, Holzinger A. Jeanquartier F, et al. BMC Bioinformatics. 2015 Jun 16;16(1):195. doi: 10.1186/s12859-015-0615-z. BMC Bioinformatics. 2015. PMID: 26077899 Free PMC article. - Bioinformatics toolbox for exploring protein phosphorylation network.
Shi XX, Wu FX, Mei LC, Wang YL, Hao GF, Yang GF. Shi XX, et al. Brief Bioinform. 2021 May 20;22(3):bbaa134. doi: 10.1093/bib/bbaa134. Brief Bioinform. 2021. PMID: 32666116 Review. - Biomolecule and Bioentity Interaction Databases in Systems Biology: A Comprehensive Review.
Baltoumas FA, Zafeiropoulou S, Karatzas E, Koutrouli M, Thanati F, Voutsadaki K, Gkonta M, Hotova J, Kasionis I, Hatzis P, Pavlopoulos GA. Baltoumas FA, et al. Biomolecules. 2021 Aug 20;11(8):1245. doi: 10.3390/biom11081245. Biomolecules. 2021. PMID: 34439912 Free PMC article. Review.
Cited by
- Patterns of HIV-1 protein interaction identify perturbed host-cellular subsystems.
MacPherson JI, Dickerson JE, Pinney JW, Robertson DL. MacPherson JI, et al. PLoS Comput Biol. 2010 Jul 29;6(7):e1000863. doi: 10.1371/journal.pcbi.1000863. PLoS Comput Biol. 2010. PMID: 20686668 Free PMC article. - Network analyses in systems pharmacology.
Berger SI, Iyengar R. Berger SI, et al. Bioinformatics. 2009 Oct 1;25(19):2466-72. doi: 10.1093/bioinformatics/btp465. Epub 2009 Jul 30. Bioinformatics. 2009. PMID: 19648136 Free PMC article. Review. - Network controllability analysis of intracellular signalling reveals viruses are actively controlling molecular systems.
Ravindran V, Nacher JC, Akutsu T, Ishitsuka M, Osadcenco A, Sunitha V, Bagler G, Schwartz JM, Robertson DL. Ravindran V, et al. Sci Rep. 2019 Feb 14;9(1):2066. doi: 10.1038/s41598-018-38224-9. Sci Rep. 2019. PMID: 30765882 Free PMC article. - GPS-Prot: a web-based visualization platform for integrating host-pathogen interaction data.
Fahey ME, Bennett MJ, Mahon C, Jäger S, Pache L, Kumar D, Shapiro A, Rao K, Chanda SK, Craik CS, Frankel AD, Krogan NJ. Fahey ME, et al. BMC Bioinformatics. 2011 Jul 22;12:298. doi: 10.1186/1471-2105-12-298. BMC Bioinformatics. 2011. PMID: 21777475 Free PMC article. - Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review.
Csermely P, Korcsmáros T, Kiss HJ, London G, Nussinov R. Csermely P, et al. Pharmacol Ther. 2013 Jun;138(3):333-408. doi: 10.1016/j.pharmthera.2013.01.016. Epub 2013 Feb 4. Pharmacol Ther. 2013. PMID: 23384594 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials