Nuclear hormone receptor regulation of microRNAs controls developmental progression - PubMed (original) (raw)
Nuclear hormone receptor regulation of microRNAs controls developmental progression
Axel Bethke et al. Science. 2009.
Abstract
In response to small-molecule signals such as retinoids or steroids, nuclear receptors activate gene expression to regulate development in different tissues. MicroRNAs turn off target gene expression within cells by binding complementary regions in messenger RNA transcripts, and they have been broadly implicated in development and disease. Here we show that the Caenorhabditis elegans nuclear receptor DAF-12 and its steroidal ligand directly activate promoters of let-7 microRNA family members to down-regulate the microRNA target hbl-1, which drives progression of epidermal stem cells from second to third larval stage patterns of cell division. Conversely, the receptor without the ligand represses microRNA expression during developmental arrest. These findings identify microRNAs as components of a hormone-coupled molecular switch that shuts off earlier developmental programs to allow for later ones.
Figures
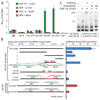
Figure 1. DAF-12 and dafachronic acid (Δ4–DA) activate microRNA promoters in vitro
A. Activation of miR promoters in HEK293T cells. Promoters of let-7 homologs, mir-84 and mir-241, are strongly activated in the presence of DAF-12 and 400nM Δ4–DA, whereas other microRNAs are relatively unaffected. Luciferase assays were measured in triplicate, (with SD). EtOH, ethanol vehicle control; ptk-luc, empty luciferase vector. B. Mutation analysis of mir-241 and mir-84 promoters reveals DAF-12/Δ4–DA activating elements. Deletion analysis of the mir-241p shows that the highest relative induction occurs with fragment #4, which contains four DAF-12 REs, 241abcd. Deletion or point mutation of 241ab elements (in red) abolishes activation (blue bars). Similarly, point mutation of DAF-12 REs in the mir-84 promoter, _mir-84_a and _mir-84_b reduce expression (red bars). C. Gel mobility shift assay of DAF-12 and mir-241p. 32P-radiolabeled oligos containing the WT 241b element are shifted (s) by nuclear extracts expressing DAF-12::FLAG and supershifted (ss) in the presence of anti-FLAG antibody. Unlabeled WT 241b-oligo competes away the shift but addition of point-mutated oligo does not.
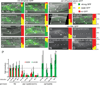
Figure 2. DAF-12 and Δ4–DA regulate microRNA promoters in vivo
Panels A–K, mir-241p::GFP. Images show representative L3 animals, with indicated cell types (white arrowhead, excretory cell, exc; outlined arrowheads, neuron, neu; muscle, mus; intestine, int,; pharynx, ph). Bar graphs quantify the percentage of worms with excretory cell GFP expression as either strong (green), weak (yellow), or off (red), (two independent experiments, left and right, n=10 animals each). mir-241p::GFP expression level in WT (N2) grown without ligand (EtOH) (A) is decreased in daf-12(rh61rh411)/NHR null (C), strongly repressed in daf-9(dh6)/CYP450 null (E) or not activated in daf-9(dh6)/CYP450;din-1(dh149)/SHARP double null (G) but rescued nearly to WT level by growth on 250 nM Δ4–DA (F, H). Δ4–DA has no effect in WT (B) or daf-12 null (D) animals. Point mutation of all four DAF-12 REs abolishes differences of tested genetic backgrounds or ligand (I–K, and Fig S1). Panels L–O, mir-84p::GFP. Epidermal seam cells (arrowheads) express mir-84p::GFP in WT N2 (L), but not in daf-12 nulls (M). Seam cell expression is absent in hormone deficient daf-9;din-1 animals (N), but restored to nearly WT levels by Δ4–DA supplementation (O). Left, percentage of L3 animals showing weak or no seam cell expression (2 independent experiments, n=20 animals each). (P) Relative quantification of microRNAs by Q-PCR. miR expression is decreased in daf-12, daf-9;din-1, and repressed in daf-9 mutants. In daf-9 genotypes, expression was Δ4–DA dependent. Q-PCR was carried out using the TaqMan system (see Fig S5 and SOM for data analysis).
Figure 3. microRNA regulation by dauer signaling pathways
mir-241p::GFP shows high expression in continuously growing WT (A), but low expression in daf-7(e1372) dauer larvae (B). mir-84p::GFP shows high expression in the pharynx of continuously growing WT (C) but low expression in daf-2(e1368) dauers (D). mir-84p::GFP seam cell expression (E) is elevated in daf-2 and daf-9 mutants during dauer stage (F, H) and is even higher in daf-7 mutants during reproductive growth at 20°C (G). Penetrant seam expression is reversed by 500 nM Δ4–DA in daf-7 but not daf-2 during reproductive growth (I–M). Animals were assayed during L3/L3d stages, n>20. Red bars, ethanol vehicle, green bars, Δ4–DA (SEM).
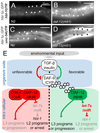
Figure 4. let-7s repression target, hbl-1, is regulated by DAF-12
A GFP-fusion to hbl-1 promoter and 3’-UTR is repressed in hypodermis at mid L3 (28h) in WT (A). In the daf-12(rh61) mutant, reporter signal is upregulated in the hypodermis (arrows) and other tissues (B) (exposure 250ms). A GFP-fusion to the hbl-1 promoter, containing the _unc-54_-3’-UTR lacks substantial up-regulation in the hypodermis (C, D), though body muscles show modest reporter upregulation (D) (exposure time 50ms) Model for nuclear receptor/microRNA signaling cascades (E). In response to favorable environmental signals, activated insulin/IGF and TGF-β pathways induce Δ4–DA biosynthesis through DAF-9/CYP450. Right: Liganded DAF-12 activates L3 programs and expression of let-7s and thereby inhibits HBL-1 and genes of L2 programs, resulting in developmental progression (E). Left: During unfavorable conditions, unliganded DAF-12 together with DIN-1, repress L3 programs and let-7s, allowing derepression of L2 programs and/or developmental arrest. Dauer signaling also has Δ4–DA-independent outputs onto miRs.
Similar articles
- miRNA regulation through ligand occupancy of a nuclear hormone receptor.
Rougvie AE. Rougvie AE. Sci Signal. 2009 Aug 18;2(84):pe52. doi: 10.1126/scisignal.284pe52. Sci Signal. 2009. PMID: 19690329 - The expression of the Alzheimer's amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans.
Niwa R, Zhou F, Li C, Slack FJ. Niwa R, et al. Dev Biol. 2008 Mar 15;315(2):418-25. doi: 10.1016/j.ydbio.2007.12.044. Epub 2008 Jan 8. Dev Biol. 2008. PMID: 18262516 Free PMC article. - The tiny RNA world.
Ruvkun GB. Ruvkun GB. Harvey Lect. 2003-2004;99:1-21. Harvey Lect. 2003. PMID: 15984549 Review. No abstract available. - Roles of microRNAs in the Caenorhabditis elegans nervous system.
Meng L, Chen L, Li Z, Wu ZX, Shan G. Meng L, et al. J Genet Genomics. 2013 Sep 20;40(9):445-52. doi: 10.1016/j.jgg.2013.07.002. Epub 2013 Aug 7. J Genet Genomics. 2013. PMID: 24053946 Review.
Cited by
- Dafachronic acid inhibits C. elegans germ cell proliferation in a DAF-12-dependent manner.
Mukherjee M, Chaudhari SN, Balachandran RS, Vagasi AS, Kipreos ET. Mukherjee M, et al. Dev Biol. 2017 Dec 15;432(2):215-221. doi: 10.1016/j.ydbio.2017.10.014. Epub 2017 Oct 21. Dev Biol. 2017. PMID: 29066181 Free PMC article. - Pheromones and Nutritional Signals Regulate the Developmental Reliance on let-7 Family MicroRNAs in C. elegans.
Ilbay O, Ambros V. Ilbay O, et al. Curr Biol. 2019 Jun 3;29(11):1735-1745.e4. doi: 10.1016/j.cub.2019.04.034. Epub 2019 May 16. Curr Biol. 2019. PMID: 31104929 Free PMC article. - cGMP and NHR signaling co-regulate expression of insulin-like peptides and developmental activation of infective larvae in Strongyloides stercoralis.
Stoltzfus JD, Bart SM, Lok JB. Stoltzfus JD, et al. PLoS Pathog. 2014 Jul 10;10(7):e1004235. doi: 10.1371/journal.ppat.1004235. eCollection 2014 Jul. PLoS Pathog. 2014. PMID: 25010340 Free PMC article. - A circadian-like gene network programs the timing and dosage of heterochronic miRNA transcription during C. elegans development.
Kinney B, Sahu S, Stec N, Hills-Muckey K, Adams DW, Wang J, Jaremko M, Joshua-Tor L, Keil W, Hammell CM. Kinney B, et al. Dev Cell. 2023 Nov 20;58(22):2563-2579.e8. doi: 10.1016/j.devcel.2023.08.006. Epub 2023 Aug 28. Dev Cell. 2023. PMID: 37643611 Free PMC article. - The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling.
Rüegger S, Miki TS, Hess D, Großhans H. Rüegger S, et al. Nucleic Acids Res. 2015 Mar 31;43(6):3344-57. doi: 10.1093/nar/gkv196. Epub 2015 Mar 9. Nucleic Acids Res. 2015. PMID: 25753661 Free PMC article.
References
- Antebi A, Culotti JG, Hedgecock EM. Development. 1998;125:1191. - PubMed
- Motola DL, et al. Cell. 2006;124:1209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM077201/GM/NIGMS NIH HHS/United States
- R01 GM077201-03/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM077201/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous