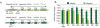The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis - PubMed (original) (raw)
The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis
Stefan Glaser et al. Epigenetics Chromatin. 2009.
Abstract
Background: Histone methylation is thought to be central to the epigenetic mechanisms that maintain and confine cellular identity in multi-cellular organisms. To examine epigenetic roles in cellular homeostasis, we conditionally mutated the histone 3 lysine 4 methyltransferase, Mll2, in embryonic stem (ES) cells, during development and in adult mice using tamoxifen-induced Cre recombination.
Results: In ES cells, expression profiling unexpectedly revealed that only one gene, Magoh2, is dependent upon Mll2 and few other genes were affected. Loss of Mll2 caused loss of H3K4me3 at the Magoh2 promoter and concomitant gain of H3K27me3 and DNA methylation. Hence Mll2, which is orthologous to Drosophila Trithorax, is required to prevent Polycomb-Group repression of the Magoh2 promoter, and repression is further accompanied by DNA methylation. Early loss of Mll2 in utero recapitulated the embryonic lethality found in Mll2-/- embryos. However, loss of Mll2 after E11.5 produced mice without notable pathologies. Hence Mll2 is not required for late development, stem cells or homeostasis in somatic cell types. However it is required in the germ cell lineage. Spermatogenesis was lost upon removal of Mll2, although spermatogonia A persisted.
Conclusion: These data suggest a bimodal recruit and maintain model whereby Mll2 is required to establish certain epigenetic decisions during differentiation, which are then maintained by redundant mechanisms. We also suggest that these mechanisms relate to the epigenetic maintenance of CpG island promoters.
Figures
Figure 1
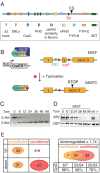
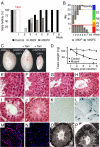
Conditional Mutagenesis of Mll2. (A) Scheme of the Mll2 protein, which contains several domains and motifs including AT hooks; SNLs (speckled nuclear localization sequences); the CxxC DNA binding region; three PHD fingers and an extended PHD finger (ePHD); a sequence similar to a Bromo domain; the FYR-N and -C domains (which dimerize); a transactivation domain (TD); a cleavage site (CS) for Taspase; and the SET domain, which is the H3K4 methyltransferase domain. (B) Scheme of the strategy for conditional mutagenesis. The CreERT2 protein was ubiquitously expressed from the Rosa26 locus but repressed by the Hsp90 complex. Tamoxifen binding releases CreERT2 from Hsp90 to permit Cre recombination of the loxP sites surrounding exon 2 of Mll2, which causes a frame-shift mutation. (C) A Southern blot to determine recombination efficiency in Mll2F/F; Rosa26-CreERT2/+ ES cells at various timepoints (0 to 96 hours) after addition of 4-hydroxy tamoxifen. (D) The same time course as shown in (C) was evaluated for Mll2 protein levels by western blotting. Wild type (wt) and Mll2-/- ES cells served as controls. (E) Summary of microarray expression profiling from Mll2-/- (constitutive) cells compared with wt or FLP rescued cells and tamoxifen-treated (conditional) compared with untreated mll2F/F; Rosa26-CreERT2/+ cells. At the left, the summary shows the number of genes whose mRNA expression level increased or decreased at least 1.7-fold in either the constitutive or conditional experiment. At the right, the overlap between the down-regulated genes in the two experiments is shown, along with the number of CpG island promoters in the three categories.
Figure 2
The Magoh2 promoter is a direct target for Mll2 and a model example of trxG/PcG opposition (part 1). (A) Diagrams of the Magoh2 and Magoh genes, showing the exon structures, the positions of the CpG island promoters (as hatched lines around exon 1) and the PCR primer pairs for chromatin immunoprecipitation (ChIP) analysis. (B) Histogram of quantitative RT-PCR analysis of Magoh (dark green) and Magoh2 (light green) mRNA levels before and after addition of 4-hydroxy tamoxifen to Mll2F/F; Rosa26-CreERT2/+ cells.
Figure 3
The Magoh2 promoter is a direct target for Mll2 and a model example of trxG/PcG opposition (part 2). ChIP analysis of the Magoh2 and Magoh promoters, as indicated at the top for the Magoh2 up, exon 1, down and Magoh exon 1 Q-PCR primer pairs as diagrammed in (Figure 2A). The first row of panels shows ChIP results obtained from TAP-tagged Mll2 ES cells using the TAP tag for immunoprecipitation compared with 5% of the input before immunoprecipitation. Foldness of Mll2-specific enrichment is noted at the side of each of the four panels. The second, third and fourth row of panels show fold change of chromatin immunoprecipitation using H3K4me3, H3K9me3 and H3K27me3-specific antibodies. Mll2F/F; Rosa26-CreERT2/+ (Mll2FC/FC)or Mll2F/+; Rosa26-CreERT2/+ (Mll2FC/+) cells were cultured for 12 days without (-) or with 4 days of 4-hydroxy tamoxifen for the last 4 days (d4), the middle 4 days (d8) or the first 4 days (d12).
Figure 4
The Magoh2 promoter is a direct target for Mll2 and a model example of trxG/PcG opposition (part 3). (A) Bisulfite sequencing results of the Magoh2 promoter before (-) and after (4-OHT) addition of tamoxifen for 4 days, followed by a further 8 days of culture shown below a diagram of the promoter, which illustrates the location of the CpG dinucleotides as well as the relevant restriction sites, Southern probe and PCR primers (small arrows either side of the Hpa1 sites) for the results shown in panels (B) and (C). (B) Southern blot to evaluate CpG methylation at the _Nae_I site of the Magoh2 promoter as illustrated in panel (A). Because the _Nae_I site contains two CpGs, resistance to restriction can reflect methylation at either or both sites, as illustrated at the right of the panel. The time course was performed on Mll2F/F; Rosa26-CreERT)/+ cells starting with addition of 4-hydroxy tamoxifen. (C) As for panel (B) except the genomic DNA samples were digested with _Hpa_I before Q-PCR analysis with the primer pair illustrated in panel (A). These data are plotted (red circles, lower plot), as well as quantitation of the Southern blot shown in panel (B; black circles, upper plot).
Figure 5
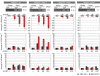
Ligand-induced mutagenesis of Mll2 in utero, via lactation and in adults. (A) Southern blot of tamoxifen-induced CreERT2 recombination in various tissues of a 2-month-old Mll2F/F; Rosa26-CreERT2/+ male two weeks after tamoxifen administration. The first lane shows tail DNA before tamoxifen administration with the 3.4 kb unrecombined band. All other lanes show DNAs after tamoxifen displaying the recombined 2.6 kb band. (B) Western blot of protein extracts from wild type and tamoxifen induced Mll2F/F; Rosa26-CreERT2/+ male testis and brain. The 300 kD protein, CBP, was used as a loading control. (C) Recombination efficiency in E10.5 embryos was detected by Southern blot. Pregnant mothers were untreated (-) or treated with one dose of 4 mg tamoxifen at E10 (12 h) or E8.5 (48 h). (D) The _Mll2_-/- phenotype was recapitulated in Mll2F/F; Rosa26-CreERT2/+ embryos after induction at E4.5 and E5.5 with doses of 1 mg tamoxifen. The embryos were harvested at E10.5. The inset shows a Mll2-/- embryo at E10.5 at the same scale. (E) Control Mll2F/+; Rosa26-CreERT2/+ embryos induced with tamoxifen at E4.5/E5.5 were normal at E10.5. (F, G) Mll2 is dispensable from E11.5. Mll2F/F; Rosa26-CreERT2/+ (F) and Mll2F/+; Rosa26-CreERT2/+ embryos (G) induced at E8.5 were normal at E12.5. (H) Southern blot of various tissues harvested from 14-day-old Mll2F/F; Rosa26-CreERT2/+ neonates, using the same strategy as in panel (A) and five daily doses of 4 mg tamoxifen to the lactating mothers, starting 4 days after delivery. (I) Body weight measurements after birth of Mll2F/F; Rosa26-CreERT2/+ or Mll2F/+; Rosa26-CreERT2/+ pups treated with tamoxifen or not as in panel (H).
Figure 6
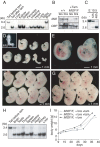
Loss of Mll2 leads to male sterility. (A) The fertility of 11 control (black) or mutant males (grey) was tested by weekly breedings to two wild-type females before and after tamoxifen administration. The mutants initially transmitted the unrecombined haplotype (light grey), and then the recombined haplotype (dark grey) and lost fertility by week 7. (B) The diagram illustrates a spermatogenic period of 40 days with mitosis (red), meiosis I (orange), meiosis II (yellow), spermiogenesis (green) and spermatozoa maturation (blue). Indicated are spermatogonia (SG), leptotene spermatocytes (LS), pachytene spematocytes (PS), rounded spermatids (RS) and elongated spermatids (ES). The transmission of the unrecombined haplotype (light grey) indicates that recombination did not occur in ES and only partially occurred in RS. Although recombination was complete in LS and PS, spermatogenesis proceeded (dark grey). Permanent sterility occurring in the sixth week indicated that spermatogenesis was interrupted before or at meiosis I (red line) (C, D) Wild-type, control and atrophic mutant testes 8 weeks after tamoxifen induction (C) or weighed during a time course (D). (E to J) Testis histological cross-sections reveal a block of germ cell differentiation and progressive loss of spermatogonia in Mll2FC/FC testis. Control testis (E) were normal after tamoxifen treatment. Mutant testis were sectioned 1 week (F), 2 weeks (G), 3 weeks (H), 4 weeks (I) and 14 weeks (J) after induction. (K, L) Increased levels of spermatogonial apoptosis in Mll2FC/FC testis, determined by TUNEL staining in control (K) and mutant (L) testis sections 3 weeks after tamoxifen induction. (M, N) Persistence of Tra98-positive cells 4 months after tamoxifen induction. Double staining with a Tra98 antibody and DAPI shows the expected distribution of Tra98 in control testis (M) and reduced staining in mutant testis (N). (O, P) Expression of Mll2 (O) and Mll (P) in normal testis detected by in situ hybridization with antisense probes.
Figure 7
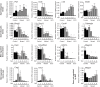
Expression analysis by quantitative RT-PCR of selected genes in mll2 mutant and control testes. The expression levels from total testes of controls (black) before (-) or 1 week after tamoxifen were compared with mutants (grey) after tamoxifen administration. All times refer to weeks after the start of five daily doses of 4 mg tamoxifen. Busulfan-treated testes were used to generate Sertoli-cell only controls (SCO). All data is based on at least three mice. The mean value ± SEM of relative expression normalized against the endogenous standard genes GAPDH and Rpl19 is shown (* P < 0.05; ** P < 0.02).
Figure 8
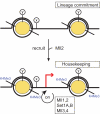
The recruit and maintain (RAM) model to explain the transient requirement for Mll2. The diagram illustrates two nucleosomes at a gene promoter, to which Mll2 is recruited. Once recruited, Mll2 mediates the establishment of a back-up system, partly via H3K4 trimethylation, which is based on its sister, Mll, and potentially the other H3K4 methyltransferase pairs, Set1A/Set1B and Mll3/Mll4. The epitopes, x and y, are included to indicate that other chromatin determinants are likely to play roles in recruitment and maintenance.
Similar articles
- Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant.
Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. Denissov S, et al. Development. 2014 Feb;141(3):526-37. doi: 10.1242/dev.102681. Epub 2014 Jan 14. Development. 2014. PMID: 24423662 - Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2.
Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Lubitz S, et al. Mol Biol Cell. 2007 Jun;18(6):2356-66. doi: 10.1091/mbc.e06-11-1060. Epub 2007 Apr 11. Mol Biol Cell. 2007. PMID: 17429066 Free PMC article. - Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation.
Douillet D, Sze CC, Ryan C, Piunti A, Shah AP, Ugarenko M, Marshall SA, Rendleman EJ, Zha D, Helmin KA, Zhao Z, Cao K, Morgan MA, Singer BD, Bartom ET, Smith ER, Shilatifard A. Douillet D, et al. Nat Genet. 2020 Jun;52(6):615-625. doi: 10.1038/s41588-020-0618-1. Epub 2020 May 11. Nat Genet. 2020. PMID: 32393859 Free PMC article. - Structure, Activity and Function of the MLL2 (KMT2B) Protein Lysine Methyltransferase.
Klonou A, Chlamydas S, Piperi C. Klonou A, et al. Life (Basel). 2021 Aug 12;11(8):823. doi: 10.3390/life11080823. Life (Basel). 2021. PMID: 34440566 Free PMC article. Review. - Mechanisms of transformation by MLL.
Hess JL. Hess JL. Crit Rev Eukaryot Gene Expr. 2004;14(4):235-54. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.10. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15663355 Review.
Cited by
- Mutations in Mll2, an H3K4 methyltransferase, result in insulin resistance and impaired glucose tolerance in mice.
Goldsworthy M, Absalom NL, Schröter D, Matthews HC, Bogani D, Moir L, Long A, Church C, Hugill A, Anstee QM, Goldin R, Thursz M, Hollfelder F, Cox RD. Goldsworthy M, et al. PLoS One. 2013 Jun 24;8(6):e61870. doi: 10.1371/journal.pone.0061870. Print 2013. PLoS One. 2013. PMID: 23826075 Free PMC article. - MLL1 is essential for retinal neurogenesis and horizontal inner neuron integrity.
Brightman DS, Grant RL, Ruzycki PA, Suzuki R, Hennig AK, Chen S. Brightman DS, et al. Sci Rep. 2018 Aug 9;8(1):11902. doi: 10.1038/s41598-018-30355-3. Sci Rep. 2018. PMID: 30093671 Free PMC article. - Histone-methyltransferase MLL2 (KMT2B) is required for memory formation in mice.
Kerimoglu C, Agis-Balboa RC, Kranz A, Stilling R, Bahari-Javan S, Benito-Garagorri E, Halder R, Burkhardt S, Stewart AF, Fischer A. Kerimoglu C, et al. J Neurosci. 2013 Feb 20;33(8):3452-64. doi: 10.1523/JNEUROSCI.3356-12.2013. J Neurosci. 2013. PMID: 23426673 Free PMC article. - Photoperiod influences growth and mll (mixed-lineage leukaemia) expression in Atlantic cod.
Nagasawa K, Giannetto A, Fernandes JM. Nagasawa K, et al. PLoS One. 2012;7(5):e36908. doi: 10.1371/journal.pone.0036908. Epub 2012 May 9. PLoS One. 2012. PMID: 22590633 Free PMC article. - Cardiac-specific inducible and conditional gene targeting in mice.
Doetschman T, Azhar M. Doetschman T, et al. Circ Res. 2012 May 25;110(11):1498-512. doi: 10.1161/CIRCRESAHA.112.265066. Circ Res. 2012. PMID: 22628574 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous