RNA polymerase V transcription guides ARGONAUTE4 to chromatin - PubMed (original) (raw)
RNA polymerase V transcription guides ARGONAUTE4 to chromatin
Andrzej T Wierzbicki et al. Nat Genet. 2009 May.
Abstract
Retrotransposons and repetitive DNA elements in eukaryotes are silenced by small RNA-directed heterochromatin formation. In Arabidopsis, this process involves 24-nt siRNAs that bind to ARGONAUTE4 (AGO4) and facilitate the targeting of complementary loci via unknown mechanisms. Nuclear RNA polymerase V (Pol V) is an RNA silencing enzyme recently shown to generate noncoding transcripts at loci silenced by 24-nt siRNAs. We show that AGO4 physically interacts with these Pol V transcripts and is thereby recruited to the corresponding chromatin. We further show that DEFECTIVE IN MERISTEM SILENCING3 (DMS3), a structural maintenance of chromosomes (SMC) hinge-domain protein, functions in the assembly of Pol V transcription initiation or elongation complexes. Collectively, our data suggest that AGO4 is guided to target loci through base-pairing of associated siRNAs with nascent Pol V transcripts.
Figures
Figure 1. Pol V, AGO4 and DMS3 work non-redundantly in heterochromatin formation
(A, B) DNA methylation analysis at the AtSN1, IGN5 and solo LTR loci in nrpe1, ago4 and dms3 mutants. Genomic DNA was digested with _Hae_III (A) or _Alu_I (B) methylation-sensitive restriction endonucleases followed by PCR. Sequences lacking _Hae_III sites (actin 2; panel A) or _Alu_I sites (IGN5, panel B) served as controls to show that equivalent amounts of DNA were tested in all reactions. (C–D) ChIP analysis of H3K27me1 (C) and H3Ac2 (D) levels in nrpe1, ago4 and dms3 mutants. Histograms show means +/− standard deviations obtained from three independent amplifications. (E) ChIP analysis of Pol II binding to chromatin in nrpe1, ago4 and dms3 mutants. Histograms show means +/− standard deviations obtained from three independent amplifications. (F) Control ChIP reactions performed in the absence of antibody reveal background signal levels.
Figure 2. AGO4 is not required for Pol V transcription
(A) Strand specific RT-PCR of Pol V transcription at IGN5 and IGN6 in ago4 and rdr2 mutants as well as nrpe1 ago4, and rdr2 ago4 double mutants. Wt sibling is a wild type sibling of the ago4 mutant identified in a segregating family. Actin RT-PCR products and ethidium bromide-stained rRNAs resolved by agarose gel electrophoresis serve as loading controls. To control for background DNA contamination, a reaction using IGN5 top strand primers but no reverse transcriptase (no RT) was performed. No RNA (0 μg) controls are provided for all primer pairs. (B) Densitometric analysis of RT-PCR data for the ago4 mutant presented in panel A. The histogram provides mean band intensities relative to wild type Col-0, +/− standard deviations obtained from three independent experiments.
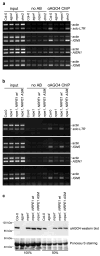
Figure 3. Pol V transcription is necessary for AGO4-chromatin interactions
(A) ChIP data showing AGO4 binding to chromatin at solo LTR, IGN5, AtSN1 and IGN6 loci in ago4, nrpe1 and drm2 mutants. DNA purified from input chromatin samples, chromatin subjected to the immunoprecipitation procedure in the absence of antibody (no AB) and chromatin immunoprecipitated using anti-AGO4 antibody (αAGO4) was amplified by PCR using locus-specific primers. Primers amplifying the Actin 2 locus served as an internal control. (B) ChIP data showing AGO4 binding to chromatin at solo LTR, IGN5, AtSN1 and IGN6 loci in nrpe1 mutant, nrpe1 mutant transformed with a wild type NRPE1 transgene (NRPE1 wt) and nrpe1 mutant transformed with an NRPE1 active site mutant transgene (NRPE1 ASM). (C) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0), ago4, nrpe1, or nrpe1 transformed with either a wild type NRPE1 transgene (NRPE1 wt) or an NRPE1 active site mutant transgene (NRPE1 ASM). Ponceau S staining revealed equal loading of lanes. 100% and 50% sample loadings demonstrate that the assay is semi-quantitative.
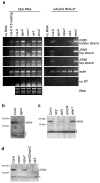
Figure 4. AGO4 physically interacts with Pol V transcripts
(A) RNA immunoprecipitation using αAGO4 antibody. Immunoprecipitated RNA isolated from the indicated mutants was digested with DNaseI and amplified by RT-PCR. Total RNA controls show that the Pol V transcripts are present in equivalent amounts in all mutants tested except nrpe1. Ethidium bromide stained rRNAs (bottom left) show that equal amounts of RNA were tested. The no reverse transcriptase (no RT) control was performed with IGN5 bottom strand primers. No RNA controls were performed for all primer pairs tested. RT-PCR amplification of actin RNA serves as a loading control. (B) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0) plants or ago4 mutant. Stars denote nonspecific bands. (C) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0), rdr2, dcl3, dcl234 or nrpe1 mutants. Asterisks denote nonspecific bands. (D) Immunoblot detection of AGO4 in protein extracts of wild-type (Col-0), nrpd1 (Pol IV) nrpe1 (Pol V), nrpd2/nrpe2 (shared subunit of Pol IV and Pol V), or rdr2 mutants. Asterisks denote nonspecific bands.
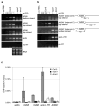
Figure 5. The SMC hinge-domain protein, DMS3 is required for Pol V transcription and detectable Pol V-chromatin interactions
(A) Strand-specific RT-PCR detection of Pol V transcripts at IGN5 and IGN6 and AtSN1 in wild-type (Col-0) and nrpe1 and dms3 mutants. Derepression of Pol II transcripts at the solo LTR and putative Pol III transcripts at AtSN1 in the nrpe1 and dms3 mutants is shown in the right panel. Actin RT-PCR products and ethidium bromide-stained rRNAs resolved by agarose gel electrophoresis serve as loading controls. To control for background DNA contamination, a reaction using IGN5 bottom strand and AtSN1 (interval B) primers, but no reverse transcriptase (no RT) was performed. No RNA (0 μg) controls are provided for all primer pairs. (B) ChIP with anti-NRPE1 antibody in Col-0 wild-type, nrpe1 and dms3 mutants followed by real-time PCR. Histograms show means +/− standard deviations obtained from three independent amplifications.
Figure 6
A model for Pol V and siRNA-dependent heterochromatin formation. DMS3 and DRD1 mediate the assembly of Pol V initiation and/or elongation complexes and the production of Pol V transcripts. AGO4-siRNA complexes recognize target loci via base-pairing of siRNAs with nascent Pol V transcripts. AGO4 subsequently recruits chromatin modifying activities including the de novo DNA methyltransferase DRM2 and histone modifying enzymes via unknown mechanisms.
Similar articles
- The ability to form homodimers is essential for RDM1 to function in RNA-directed DNA methylation.
Sasaki T, Lorković ZJ, Liang SC, Matzke AJ, Matzke M. Sasaki T, et al. PLoS One. 2014 Feb 3;9(2):e88190. doi: 10.1371/journal.pone.0088190. eCollection 2014. PLoS One. 2014. PMID: 24498436 Free PMC article. - Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing.
Rowley MJ, Avrutsky MI, Sifuentes CJ, Pereira L, Wierzbicki AT. Rowley MJ, et al. PLoS Genet. 2011 Jun;7(6):e1002120. doi: 10.1371/journal.pgen.1002120. Epub 2011 Jun 9. PLoS Genet. 2011. PMID: 21738482 Free PMC article. - RNA-directed DNA methylation involves co-transcriptional small-RNA-guided slicing of polymerase V transcripts in Arabidopsis.
Liu W, Duttke SH, Hetzel J, Groth M, Feng S, Gallego-Bartolome J, Zhong Z, Kuo HY, Wang Z, Zhai J, Chory J, Jacobsen SE. Liu W, et al. Nat Plants. 2018 Mar;4(3):181-188. doi: 10.1038/s41477-017-0100-y. Epub 2018 Jan 29. Nat Plants. 2018. PMID: 29379150 Free PMC article. - Cell biology of the Arabidopsis nuclear siRNA pathway for RNA-directed chromatin modification.
Pikaard CS. Pikaard CS. Cold Spring Harb Symp Quant Biol. 2006;71:473-80. doi: 10.1101/sqb.2006.71.046. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381329 Review. - The role of long non-coding RNA in transcriptional gene silencing.
Wierzbicki AT. Wierzbicki AT. Curr Opin Plant Biol. 2012 Nov;15(5):517-22. doi: 10.1016/j.pbi.2012.08.008. Epub 2012 Sep 6. Curr Opin Plant Biol. 2012. PMID: 22960034 Review.
Cited by
- Mathematical model of RNA-directed DNA methylation predicts tuning of negative feedback required for stable maintenance.
Dale R, Mosher R. Dale R, et al. Open Biol. 2024 Nov;14(11):240159. doi: 10.1098/rsob.240159. Epub 2024 Nov 13. Open Biol. 2024. PMID: 39532148 Free PMC article. - Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review. - An RdDM-independent function of Pol V transcripts in gene regulation and plant defence.
Yuan Y, Liu Y, Han L, Li Y, Qi Y. Yuan Y, et al. Nat Plants. 2024 Oct;10(10):1562-1575. doi: 10.1038/s41477-024-01774-0. Epub 2024 Aug 26. Nat Plants. 2024. PMID: 39187700 - Transcription elongation of the plant RNA polymerase IV is prone to backtracking.
Fang C, Huang K, Wu X, Zhang H, Gu Z, Wang J, Zhang Y. Fang C, et al. Sci Adv. 2024 Aug 23;10(34):eadq3087. doi: 10.1126/sciadv.adq3087. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178250 Free PMC article. - H1 restricts euchromatin-associated methylation pathways from heterochromatic encroachment.
Harris CJ, Zhong Z, Ichino L, Feng S, Jacobsen SE. Harris CJ, et al. Elife. 2024 May 30;12:RP89353. doi: 10.7554/eLife.89353. Elife. 2024. PMID: 38814684 Free PMC article.
References
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–96. - PubMed
- Kanno T, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–5. - PubMed
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials