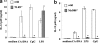Toll-like receptor 9-dependent activation of myeloid dendritic cells by Deoxynucleic acids from Candida albicans - PubMed (original) (raw)
. 2009 Jul;77(7):3056-64.
doi: 10.1128/IAI.00840-08. Epub 2009 May 11.
Kiwamu Nakamura, Natsuo Yamamoto, Héctor M Mora-Montes, Misuzu Tanaka, Yuzuru Abe, Daiki Tanno, Ken Inden, Xiao Gang, Keiko Ishii, Kiyoshi Takeda, Shizuo Akira, Shinobu Saijo, Yoichiro Iwakura, Yoshiyuki Adachi, Naohito Ohno, Kotaro Mitsutake, Neil A R Gow, Mitsuo Kaku, Kazuyoshi Kawakami
Affiliations
- PMID: 19433551
- PMCID: PMC2708591
- DOI: 10.1128/IAI.00840-08
Toll-like receptor 9-dependent activation of myeloid dendritic cells by Deoxynucleic acids from Candida albicans
Akiko Miyazato et al. Infect Immun. 2009 Jul.
Abstract
The innate immune system of humans recognizes the human pathogenic fungus Candida albicans via sugar polymers present in the cell wall, such as mannan and beta-glucan. Here, we examined whether nucleic acids from C. albicans activate dendritic cells. C. albicans DNA induced interleukin-12p40 (IL-12p40) production and CD40 expression by murine bone marrow-derived myeloid dendritic cells (BM-DCs) in a dose-dependent manner. BM-DCs that lacked Toll-like receptor 4 (TLR4), TLR2, and dectin-1, which are pattern recognition receptors for fungal cell wall components, produced IL-12p40 at levels comparable to the levels produced by BM-DCs from wild-type mice, and DNA from a C. albicans pmr1Delta null mutant, which has a gross defect in mannosylation, retained the ability to activate BM-DCs. This stimulatory effect disappeared completely after DNase treatment. In contrast, RNase treatment increased production of the cytokine. A similar reduction in cytokine production was observed when BM-DCs from TLR9(-/-) and MyD88(-/-) mice were used. In a luciferase reporter assay, NF-kappaB activation was detected in TLR9-expressing HEK293T cells stimulated with C. albicans DNA. Confocal microscopic analysis showed similar localization of C. albicans DNA and CpG-oligodeoxynucleotide (CpG-ODN) in BM-DCs. Treatment of C. albicans DNA with methylase did not affect its ability to induce IL-12p40 synthesis, whereas the same treatment completely eliminated the ability of CpG-ODN to induce IL-12p40 synthesis. Finally, impaired clearance of this fungal pathogen was not found in the kidneys of TLR9(-/-) mice. These results suggested that C. albicans DNA activated BM-DCs through a TLR9-mediated signaling pathway using a mechanism independent of the unmethylated CpG motif.
Figures
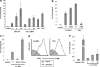
FIG. 1.
C. albicans DNA and S. cerevisiae DNA activate BM-DCs. BM-DCs were cultured under various conditions for 24 h. IL-12p40 levels in the culture supernatants were determined by ELISA (a, b, c, and e), and surface expression of CD40 on BM-DCs was determined by flow cytometry (d). The data are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments. (−) indicates C. albicans DNA (CA-DNA) stimulation without polymyxin B (b) or DNase or RNase (c) treatment. *, P < 0.05 for a comparison of a preparation treated with polymyxin B, DNase, or RNase and a preparation that was not treated. The amount of DNA from C. albicans THK519 and S. cerevisiae DNA used was 10 μg/ml, unless indicated otherwise (a and e). The concentration of LPS used was 1 μg/ml, and the concentration of DNase used was 10 μg/ml. PMB, polymyxin B (10 μg/ml); CpG, CpG1826 (1 μg/ml).
FIG. 2.
Role of C. albicans cell wall components in the activation of BM-DCs. BM-DCs from TLR4−/− and wild-type mice (a), TLR2−/− and wild-type mice (b), and dectin-1−/− and wild-type mice (c) were cultured with DNA from C. albicans THK519 (10 μg/ml), CpG1826 (1 μg/ml), LPS (1 μg/ml), OX-CA (3 μg/ml), or PG (1 μg/ml) for 24 h, and IL-12p40 levels in the culture supernatants were determined by ELISA. The data are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments. CA-DNA, C. albicans DNA; CpG, CpG1826. *, P < 0.05 for a comparison of TLR4−/−, TLR2−/−, or dectin-1−/− mice and wild-type mice.
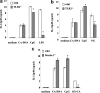
FIG. 3.
Role of manosyl residues in the activation of BM-DCs. BM-DCs were cultured with C. albicans DNA extracted from wild-type strain CAI4 or the _C. albicans pmr1_Δ null mutant at concentrations of 1, 3, and 10 μg/ml for 24 h, and IL-12p40 levels in the culture supernatants were determined by ELISA (a). C. albicans DNA was pretreated with or without DNase (b). (−) indicates C. albicans DNA stimulation without DNase treatment. *, P < 0.05 for a comparison of a preparation treated with DNase and a preparation that was not treated. The data are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments.
FIG. 4.
Inhibitory effect of _C. albicans pmr1_Δ DNA on THK DNA activity. BM-DCs were incubated with 3 μg/ml DNA from THK519 alone or with various concentrations of _pmr1_Δ null mutant DNA (50, 25, and 12.5 μg/ml) or with _pmr1_Δ DNA alone (50, 25, and 12.5 μg/ml) for 24 h. IL-12p40 levels in the culture supernatants were measured by ELISA. The data are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments. *, P < 0.05 for a comparison of a preparation incubated with THK519 alone and a preparation incubated with _pmr1_Δ null mutant DNA.
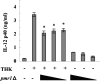
FIG. 5.
Indispensable role of TLR9-MyD88 signaling pathway in BM-DC activation. BM-DCs from TLR9−/− and wild-type mice (a) and MyD88−/− and wild-type mice (b) were cultured with C. albicans ATCC 18804 DNA (10 μg/ml), CpG (1 μg/ml), or LPS (1 μg/ml) for 24 h, and IL-12p40 levels in the culture supernatants were measured by ELISA. The data are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments. CA-DNA, C. albicans DNA; CpG, CpG1826. *, P < 0.05 for a comparison of TLR9−/− or MyD88−/− mice and wild-type mice.
FIG. 6.
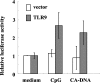
NF-κB activation via TLR9 by C. albicans DNA. HEK293T cells transfected with a TLR9 gene or control vector were cultured with CpG1826 (300 nM) or DNA from C. albicans THK519 (100 μg/ml) for 6 h. The luciferase activity in each sample was determined as described in Materials and Methods. Data are expressed as relative values based on the value for an unstimulated sample in each group and are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments. CA-DNA, C. albicans DNA; CpG, CpG1826.
FIG. 7.
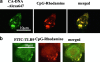
Trafficking of C. albicans DNA in BM-DCs. BM-DCs were simultaneously incubated with 10 μg/ml Alexa 647-conjugated DNA from C. albicans THK519 (green) and 3 μM CpG-rhodamine (red) (a). BM-DCs were incubated with 3 μM CpG-rhodamine (red) for 30 min. After fixation, TLR9 was stained intracellularly by direct immunofluorescence with FITC-conjugated anti-TLR9 antibody (green) (b). Cells were analyzed using a confocal microscope. The data are representative of three or four independent experiments. CA-DNA, C. albicans DNA; CpG, CpG1826.
FIG. 8.
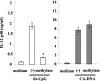
Effect of methylation of C. albicans DNA. BM-DCs were cultured with ds CpG1826 (4.5 μM) or DNA from C. albicans THK519 (10 μg/ml) pretreated or not pretreated with methylase. IL-12p40 levels in the cultures supernatants were measured by ELISA. The data are means ± SD of triplicate cultures. Similar results were obtained in three independent experiments. (−) indicates C. albicans DNA stimulation without methylase treatment. CA-DNA, C. albicans DNA; ds-CpG, ds CpG1826. *, P < 0.05 for a comparison of a preparation treated with methylase and a preparation that was not treated with methylase.
FIG. 9.
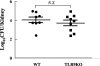
Fungal growth in TLR9−/− and wild-type mice with systemic C. albicans infections. Wild-type or TLR9−/− mice were inoculated intravenously with 105 C. albicans yeast cells in 0.1 ml. Fungal growth in the kidneys was assessed 5 days after infection. Fungal growth in the kidneys (means ± standard errors) is expressed as the number of CFU per organ. WT, wild-type mice; TLR9KO, TLR9−/− mice; N.S., difference not significant.
Similar articles
- Deoxynucleic acids from Cryptococcus neoformans activate myeloid dendritic cells via a TLR9-dependent pathway.
Nakamura K, Miyazato A, Xiao G, Hatta M, Inden K, Aoyagi T, Shiratori K, Takeda K, Akira S, Saijo S, Iwakura Y, Adachi Y, Ohno N, Suzuki K, Fujita J, Kaku M, Kawakami K. Nakamura K, et al. J Immunol. 2008 Mar 15;180(6):4067-74. doi: 10.4049/jimmunol.180.6.4067. J Immunol. 2008. PMID: 18322216 - Toll-like receptor 9-dependent activation of bone marrow-derived dendritic cells by URA5 DNA from Cryptococcus neoformans.
Tanaka M, Ishii K, Nakamura Y, Miyazato A, Maki A, Abe Y, Miyasaka T, Yamamoto H, Akahori Y, Fue M, Takahashi Y, Kanno E, Maruyama R, Kawakami K. Tanaka M, et al. Infect Immun. 2012 Feb;80(2):778-86. doi: 10.1128/IAI.05570-11. Epub 2011 Nov 21. Infect Immun. 2012. PMID: 22104112 Free PMC article. - Activation of myeloid dendritic cells by deoxynucleic acids from Cordyceps sinensis via a Toll-like receptor 9-dependent pathway.
Xiao G, Miyazato A, Abe Y, Zhang T, Nakamura K, Inden K, Tanaka M, Tanno D, Miyasaka T, Ishii K, Takeda K, Akira S, Saijo S, Iwakura Y, Adachi Y, Ohno N, Yamamoto N, Kunishima H, Hirakata Y, Kaku M, Kawakami K. Xiao G, et al. Cell Immunol. 2010;263(2):241-50. doi: 10.1016/j.cellimm.2010.04.006. Epub 2010 Apr 20. Cell Immunol. 2010. PMID: 20451901 - Toll-like receptor 4-dependent activation of myeloid dendritic cells by leukocidin of Staphylococcus aureus.
Inden K, Kaneko J, Miyazato A, Yamamoto N, Mouri S, Shibuya Y, Nakamura K, Aoyagi T, Hatta M, Kunishima H, Hirakata Y, Itoh Y, Kaku M, Kawakami K. Inden K, et al. Microbes Infect. 2009 Feb;11(2):245-53. doi: 10.1016/j.micinf.2008.11.013. Epub 2008 Dec 14. Microbes Infect. 2009. PMID: 19111627 - Cryptococcus neoformans suppresses the activation of bone marrow-derived dendritic cells stimulated with its own DNA, but not with DNA from other fungi.
Yamamoto H, Abe Y, Miyazato A, Tanno D, Tanaka M, Miyasaka T, Ishii K, Kawakami K. Yamamoto H, et al. FEMS Immunol Med Microbiol. 2011 Dec;63(3):363-72. doi: 10.1111/j.1574-695X.2011.00859.x. Epub 2011 Sep 16. FEMS Immunol Med Microbiol. 2011. PMID: 22092563
Cited by
- Interplay between Candida albicans and the mammalian innate host defense.
Cheng SC, Joosten LA, Kullberg BJ, Netea MG. Cheng SC, et al. Infect Immun. 2012 Apr;80(4):1304-13. doi: 10.1128/IAI.06146-11. Epub 2012 Jan 17. Infect Immun. 2012. PMID: 22252867 Free PMC article. Review. - LncRNA: A Potential Target for Host-Directed Therapy of Candida Infection.
Wang Y, Xu H, Chen N, Yang J, Zhou H. Wang Y, et al. Pharmaceutics. 2022 Mar 11;14(3):621. doi: 10.3390/pharmaceutics14030621. Pharmaceutics. 2022. PMID: 35335994 Free PMC article. Review. - The cell biology of the innate immune response to Aspergillus fumigatus.
Mansour MK, Tam JM, Vyas JM. Mansour MK, et al. Ann N Y Acad Sci. 2012 Dec;1273(1):78-84. doi: 10.1111/j.1749-6632.2012.06837.x. Ann N Y Acad Sci. 2012. PMID: 23230841 Free PMC article. - Mannosylation in Candida albicans: role in cell wall function and immune recognition.
Hall RA, Gow NA. Hall RA, et al. Mol Microbiol. 2013 Dec;90(6):1147-61. doi: 10.1111/mmi.12426. Epub 2013 Nov 8. Mol Microbiol. 2013. PMID: 24125554 Free PMC article. Review. - Innate immune cell response upon Candida albicans infection.
Qin Y, Zhang L, Xu Z, Zhang J, Jiang YY, Cao Y, Yan T. Qin Y, et al. Virulence. 2016 Jul 3;7(5):512-26. doi: 10.1080/21505594.2016.1138201. Epub 2016 Apr 14. Virulence. 2016. PMID: 27078171 Free PMC article. Review.
References
- Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. - PubMed
- Bates, S., M. D. MacCallum, G. Bertram, C. A. Munro, H. B. Hughes, E. T. Buurman, A. J. P. Brown, F. C. Odds, and N. A. R. Gow. 2005. Candida albicans Pmr1p, a secretory pathway P-type Ca2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 28023408-23415. - PubMed
- Bellocchio, S., C. Montagnoli, S. Bozza, R. Gaziano, G. Rossi, S. S. Mambula, A. Vecchi, A. Mantovani, S. M. Levitz, and L. Romani. 2004. The contribution of the Toll-like/IL-1 receptor superfamily to innate and adaptive immunity to fungal pathogens in vivo. J. Immunol. 1723059-3069. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials