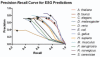ESG: extended similarity group method for automated protein function prediction - PubMed (original) (raw)
ESG: extended similarity group method for automated protein function prediction
Meghana Chitale et al. Bioinformatics. 2009.
Abstract
Motivation: Importance of accurate automatic protein function prediction is ever increasing in the face of a large number of newly sequenced genomes and proteomics data that are awaiting biological interpretation. Conventional methods have focused on high sequence similarity-based annotation transfer which relies on the concept of homology. However, many cases have been reported that simple transfer of function from top hits of a homology search causes erroneous annotation. New methods are required to handle the sequence similarity in a more robust way to combine together signals from strongly and weakly similar proteins for effectively predicting function for unknown proteins with high reliability.
Results: We present the extended similarity group (ESG) method, which performs iterative sequence database searches and annotates a query sequence with Gene Ontology terms. Each annotation is assigned with probability based on its relative similarity score with the multiple-level neighbors in the protein similarity graph. We will depict how the statistical framework of ESG improves the prediction accuracy by iteratively taking into account the neighborhood of query protein in the sequence similarity space. ESG outperforms conventional PSI-BLAST and the protein function prediction (PFP) algorithm. It is found that the iterative search is effective in capturing multiple-domains in a query protein, enabling accurately predicting several functions which originate from different domains.
Availability: ESG web server is available for automated protein function prediction at http://dragon.bio.purdue.edu/ESG/.
Figures
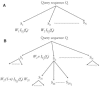
Fig. 1.
(A) ESG computation with one level. Sequences _S_1 to S N are retrieved by PSI-BLAST search. (B) ESG with two levels. The second round of PSI-BLAST searches are performed from each of the sequences, _S_1 to S N.
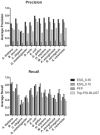
Fig. 2.
Precision–recall curve of ESG predictions. Crosses show the data points with the probability cutoff of 0.35.
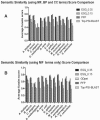
Fig. 3.
Prediction accuracy measured by the funsim score. (A) Average funsim scores for ESG, PFP and Top PSI-BLAST for benchmarking dataset across 12 species. (B), Comparison with GOPET with funsim score using MF terms only.
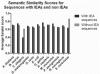
Fig. 4.
The funsim score for benchmarking set with and without IEAs across the 12 different organisms. Probability cutoff of 0.35 is used.
Fig. 5.
Precision and recall values for ESG, PFP and Top PSI-BLAST.
Fig. 6.
Domain structure of PDGFRB, PRKG1 and NCAM2 (figure is not drawn to the exact scale of the proteins).
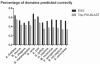
Fig. 7.
Domain assignment accuracy.
Fig. 8.
Heatmap representation of sequence hits by ESG for a query sequence, P76216. The left most column shows sequence hits by the first-level ESG search sorted by the _E_-value. Each row represents one sequence. The top 50 sequences used as queries in the second-level search are surrounded by a thick rectangle. Sequences below the thick rectangle have an _E_-value between 10 and 13. The second-level search results from each of the 50 sequences are visualized in the next 50 columns. In columns of the second-level search, gray boxes indicate that the sequences found in the first-level search reappeared in the second-level search. Black boxes at the right side of each row indicate that the sequence representing the row has annotation common with the query. The figure represents the top part of sequences obtained in the ESG computation.
Similar articles
- In-depth performance evaluation of PFP and ESG sequence-based function prediction methods in CAFA 2011 experiment.
Chitale M, Khan IK, Kihara D. Chitale M, et al. BMC Bioinformatics. 2013;14 Suppl 3(Suppl 3):S2. doi: 10.1186/1471-2105-14-S3-S2. Epub 2013 Feb 28. BMC Bioinformatics. 2013. PMID: 23514353 Free PMC article. - Using PFP and ESG Protein Function Prediction Web Servers.
Wei Q, McGraw J, Khan I, Kihara D. Wei Q, et al. Methods Mol Biol. 2017;1611:1-14. doi: 10.1007/978-1-4939-7015-5_1. Methods Mol Biol. 2017. PMID: 28451967 - PFP/ESG: automated protein function prediction servers enhanced with Gene Ontology visualization tool.
Khan IK, Wei Q, Chitale M, Kihara D. Khan IK, et al. Bioinformatics. 2015 Jan 15;31(2):271-2. doi: 10.1093/bioinformatics/btu646. Epub 2014 Oct 1. Bioinformatics. 2015. PMID: 25273111 Free PMC article. - Gene Ontology annotation of the rice blast fungus, Magnaporthe oryzae.
Meng S, Brown DE, Ebbole DJ, Torto-Alalibo T, Oh YY, Deng J, Mitchell TK, Dean RA. Meng S, et al. BMC Microbiol. 2009 Feb 19;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2180-9-S1-S8. BMC Microbiol. 2009. PMID: 19278556 Free PMC article. Review. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
- In-depth performance evaluation of PFP and ESG sequence-based function prediction methods in CAFA 2011 experiment.
Chitale M, Khan IK, Kihara D. Chitale M, et al. BMC Bioinformatics. 2013;14 Suppl 3(Suppl 3):S2. doi: 10.1186/1471-2105-14-S3-S2. Epub 2013 Feb 28. BMC Bioinformatics. 2013. PMID: 23514353 Free PMC article. - Assessing the Performances of Protein Function Prediction Algorithms from the Perspectives of Identification Accuracy and False Discovery Rate.
Yu CY, Li XX, Yang H, Li YH, Xue WW, Chen YZ, Tao L, Zhu F. Yu CY, et al. Int J Mol Sci. 2018 Jan 8;19(1):183. doi: 10.3390/ijms19010183. Int J Mol Sci. 2018. PMID: 29316706 Free PMC article. - EnzymeDetector: an integrated enzyme function prediction tool and database.
Quester S, Schomburg D. Quester S, et al. BMC Bioinformatics. 2011 Sep 23;12:376. doi: 10.1186/1471-2105-12-376. BMC Bioinformatics. 2011. PMID: 21943292 Free PMC article. - The PFP and ESG protein function prediction methods in 2014: effect of database updates and ensemble approaches.
Khan IK, Wei Q, Chapman S, Kc DB, Kihara D. Khan IK, et al. Gigascience. 2015 Sep 14;4:43. doi: 10.1186/s13742-015-0083-4. eCollection 2015. Gigascience. 2015. PMID: 26380077 Free PMC article. - Chromosome level genome assembly of the Etruscan shrew Suncus etruscus.
Bukhman YV, Meyer S, Chu LF, Abueg L, Antosiewicz-Bourget J, Balacco J, Brecht M, Dinatale E, Fedrigo O, Formenti G, Fungtammasan A, Giri SJ, Hiller M, Howe K, Kihara D, Mamott D, Mountcastle J, Pelan S, Rabbani K, Sims Y, Tracey A, Wood JMD, Jarvis ED, Thomson JA, Chaisson MJP, Stewart R. Bukhman YV, et al. Sci Data. 2024 Feb 7;11(1):176. doi: 10.1038/s41597-024-03011-x. Sci Data. 2024. PMID: 38326333 Free PMC article.
References
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Devos D, Valencia A. Intrinsic errors in genome annotation. Trends Genet. 2001;17:429–431. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials