The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes - PubMed (original) (raw)
The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes
Shinpei Kawaoka et al. RNA. 2009 Jul.
Abstract
Genetic studies and large-scale sequencing experiments have revealed that the PIWI subfamily proteins and PIWI-interacting RNAs (piRNAs) play an important role in germ line development and transposon control. Biochemical studies in vitro have greatly contributed to the understanding of small interfering RNA (siRNA) and microRNA (miRNA) pathways. However, in vitro analyses of the piRNA pathway have been thus far quite challenging, because their expression is largely restricted to the germ line. Here we report that Bombyx mori ovary-derived cultured cell line, BmN4, endogenously expresses two PIWI subfamily proteins, silkworm Piwi (Siwi) and Ago3 (BmAgo3), and piRNAs associated with them. Siwi-bound piRNAs have a strong bias for uridine at their 5' end and BmAgo3-bound piRNAs are enriched for adenine at position 10. In addition, Siwi preferentially binds antisense piRNAs, whereas BmAgo3 binds sense piRNAs. Moreover, we identified many pairs in which Siwi-bound antisense and BmAgo3-bound sense piRNAs are overlapped by precisely 10 nt at their 5' ends. These signatures are known to be important for secondary piRNA biogenesis in other organisms. Taken together, BmN4 is a unique cell line in which both primary and secondary steps of piRNA biogenesis pathways are active. This cell line would provide useful tools for analysis of piRNA biogenesis and function.
Figures
FIGURE 1.
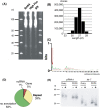
Identification and characterization of BmN4-derived small RNAs. (A) Total RNA (10 μg) from three lepidopteran cultured cell lines (BmN4, High Five, and Sf9) were loaded on a 15% polyacrylamide gel containing 10 M urea, electrophoresed, and stained with SYBRGold. Molecular sizes are shown to the left of the figure. The arrow indicates the small RNA species of interest. (B) Length distributions of sequenced BmN4-derived piRNAs. BmN4-derived piRNAs displayed a unimodal length distribution with a peak at 27 nt. (C) Nucleotide compositions of sequenced total BmN4-derived piRNAs. BmN4-derived piRNAs have a strong bias toward uridine at the 5′ end (76%). (D) Pie chart summarizing the annotation of BmN4-derived small RNAs. (E) Chemical characterization of 3′ end of Bombyx piRNA. Northern blot analysis of ovary- and BmN4-derived total RNAs after NaIO4-mediated oxidation followed by β-elimination reaction (indicated by +) was performed. Molecular sizes are shown to the left of the figure and DNA probes used are indicated above.
FIGURE 2.
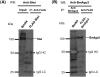
Expressions of Siwi and BmAgo3 in BmN4 cells. (A) Expression of Siwi in BmN4 cells. Western blotting was performed using total protein extract from BmN4 cells. The immunoprecipitates prepared with anti-FLAG antibody from extracts of BmN4 cells stably expressing FLAG-Siwi were used as a positive control. Siwi was expressed as an ∼100-kDa protein in BmN4 cells. (B) Expression of BmAgo3 in BmN4 cells. BmAgo3 was not detected by anti-BmAgo3 antibody using total protein lysate from BmN4 cells (data not shown). Thus, Western blotting was performed using the immunoprecipitates prepared with anti-BmAgo3 antibody from extracts of BmN4 cells. The immunoprecipitates prepared with anti-FLAG antibody from extracts of BmN4 cells stably expressing FLAG-BmAgo3 were used as a positive control. BmAgo3 was expressed as an ∼110 kDa protein in BmN4 cells.
FIGURE 3.
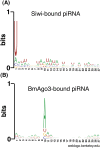
Nucleotide biases of Siwi- and BmAgo3-bound piRNAs in BmN4 cells. (A) Nucleotide bias of Siwi-bound piRNAs. (B) Nucleotide bias of BmAgo3-bound piRNAs.
FIGURE 4.
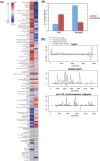
Characterization of transposon-associated Siwi- and BmAgo3-bound piRNAs (A) The heat map indicates the number of clones of Siwi- and BmAgo3-bound piRNAs that matched to transposon sequences in the Bombyx genome. Red shows antisense clones and blue shows sense clones. (B) Strand bias of Siwi- and BmAgo3-bound piRNAs. (C) Shown is the density of individual transposon-associated piRNAs. Frequency observed in the BmN4-originated piRNAs is shown individually along the y-axis. Relative nucleotide position of each transposon is indicated along the x-axis.
FIGURE 5.
The 10-nt overlap from 5′ ends between Siwi- and BmAgo3-bound piRNAs. (A) The 10- nt overlap between Siwi-bound antisense piRNAs and BmAgo3-bound sense piRNAs. We calculated the number of ping-pong “sites” within each transposons and plotted total numbers of sites at each overlapping length. The number of overlapping sites is shown along the y-axis. The overlapping length at the 5′ end is indicated along the x-axis. (B) Shown are heat maps that indicate the frequency to which 10-nt overlapping sense–antisense piRNA pairs from position 1–10 are found. Total small RNAs, SIWI-bound piRNAs, and BmAgo3-bound piRNAs were used in this analysis. Right panel indicates a control analysis performed with the 9-nt overlap.
Similar articles
- Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells.
Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T. Kawaoka S, et al. Biochem Biophys Res Commun. 2008 Mar 21;367(4):755-60. doi: 10.1016/j.bbrc.2008.01.013. Epub 2008 Jan 10. Biochem Biophys Res Commun. 2008. PMID: 18191035 - Molecular characterization of mitochondrial Zucchini and its relation to nuage-piRNA pathway components in Bombyx mori ovary-derived BmN4 cells.
Patil AA, Tatsuke T, Mon H, Lee JM, Morokuma D, Hino M, Kusakabe T. Patil AA, et al. Biochem Biophys Res Commun. 2017 Nov 18;493(2):971-978. doi: 10.1016/j.bbrc.2017.09.107. Epub 2017 Sep 20. Biochem Biophys Res Commun. 2017. PMID: 28942151 - Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells.
Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, Siomi MC. Nishida KM, et al. Cell Rep. 2015 Jan 13;10(2):193-203. doi: 10.1016/j.celrep.2014.12.013. Epub 2014 Dec 31. Cell Rep. 2015. PMID: 25558067 - The biogenesis and function of PIWI proteins and piRNAs: progress and prospect.
Thomson T, Lin H. Thomson T, et al. Annu Rev Cell Dev Biol. 2009;25:355-76. doi: 10.1146/annurev.cellbio.24.110707.175327. Annu Rev Cell Dev Biol. 2009. PMID: 19575643 Free PMC article. Review. - PIWI pathway against viruses in insects.
Kolliopoulou A, Santos D, Taning CNT, Wynant N, Vanden Broeck J, Smagghe G, Swevers L. Kolliopoulou A, et al. Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1555. doi: 10.1002/wrna.1555. Epub 2019 Jun 10. Wiley Interdiscip Rev RNA. 2019. PMID: 31183996 Review.
Cited by
- Transposable-element associated small RNAs in Bombyx mori genome.
Cai Y, Zhou Q, Yu C, Wang X, Hu S, Yu J, Yu X. Cai Y, et al. PLoS One. 2012;7(5):e36599. doi: 10.1371/journal.pone.0036599. Epub 2012 May 8. PLoS One. 2012. PMID: 22662121 Free PMC article. - The silkworm W chromosome is a source of female-enriched piRNAs.
Kawaoka S, Kadota K, Arai Y, Suzuki Y, Fujii T, Abe H, Yasukochi Y, Mita K, Sugano S, Shimizu K, Tomari Y, Shimada T, Katsuma S. Kawaoka S, et al. RNA. 2011 Dec;17(12):2144-51. doi: 10.1261/rna.027565.111. Epub 2011 Oct 21. RNA. 2011. PMID: 22020973 Free PMC article. - Silkworm HP1a transcriptionally enhances highly expressed euchromatic genes via association with their transcription start sites.
Shoji K, Hara K, Kawamoto M, Kiuchi T, Kawaoka S, Sugano S, Shimada T, Suzuki Y, Katsuma S. Shoji K, et al. Nucleic Acids Res. 2014 Oct;42(18):11462-71. doi: 10.1093/nar/gku862. Epub 2014 Sep 18. Nucleic Acids Res. 2014. PMID: 25237056 Free PMC article. - PIWI Slicing and EXD1 Drive Biogenesis of Nuclear piRNAs from Cytosolic Targets of the Mouse piRNA Pathway.
Yang Z, Chen KM, Pandey RR, Homolka D, Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA, Pillai RS. Yang Z, et al. Mol Cell. 2016 Jan 7;61(1):138-52. doi: 10.1016/j.molcel.2015.11.009. Epub 2015 Dec 6. Mol Cell. 2016. PMID: 26669262 Free PMC article. - Functional Characterization of Silkworm PIWI Proteins by Embryonic RNAi.
Kiuchi T, Katsuma S. Kiuchi T, et al. Methods Mol Biol. 2022;2360:19-31. doi: 10.1007/978-1-0716-1633-8_3. Methods Mol Biol. 2022. PMID: 34495504
References
- Bently DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16:545–552. - PubMed
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials