Phosphorylation state defines discrete roles for monopolin in chromosome attachment and spindle elongation - PubMed (original) (raw)
Phosphorylation state defines discrete roles for monopolin in chromosome attachment and spindle elongation
Sung Hugh Choi et al. Curr Biol. 2009.
Abstract
Background: It is unknown how oscillations in Cdk1 activity drive the dramatic changes in chromosome and spindle dynamics that occur at the metaphase/anaphase transition.
Results: We show that the Schizosaccharomyces pombe monopolin complex has distinct functions in metaphase and anaphase that are determined by the phosphorylation state of its Mde4 subunit. When Cdk1 activity is high in metaphase, Mde4 is hyperphosphorylated on Cdk1 phosphorylation sites and localizes to kinetochores. A nonphosphorylatable mutant of Mde4 does not localize to kinetochores, appears prematurely on the metaphase spindle, and interferes with spindle dynamics and chromosome segregation, illustrating the importance of Cdk1 phosphorylation in regulating metaphase monopolin activity. When Cdk1 activity drops in anaphase, dephosphorylation of Mde4 triggers monopolin localization to the mitotic spindle, where it promotes spindle elongation and integrity, coupling the late mitotic loss of Cdk1 activity to anaphase spindle dynamics.
Conclusions: Together, these findings illustrate how the sequential phosphorylation and dephosphorylation of monopolin helps ensure the orderly execution of discrete steps in mitosis.
Figures
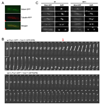
Figure 1. Mde4 is phosphorylated during mitosis and interacts with Clp1
(A) Schematic representation of Mde4 depicting 12 consensus Cdk1 phosphorylation sites. 5 of the 12 predicted phosphorylation sites shown as red ℗ were identified by mass spectrometric analysis of Clp1-C286S-TAP purified from cells arrested at metaphase using the nda3-KM311 and mts3-1 mutants and from cells 60 min after release from a cdc25-22 arrest. (B) Interaction between Mde4 and Clp1 was determined by immunoprecipitation followed by western blotting using cell lysates from the following asynchronous cultures: wild-type (1), clp1-13Myc (2), mde4-GFP (3), mde4-GFP clp1-13Myc (4), and mde4-GFP clp1-C286S-13Myc (5). The lower panel shows the tubulin loading control from whole cell extracts prior to immunoprecipitation. (C) Cell cycle dependent changes in Mde4 phosphorylation was determined by analyzing the gel migration of Mde4-13Myc. cdc25-22 mde4-13Myc cells were arrested at the restrictive temperature of 36°C for 4 hours to synchronize them in G2 phase, then shifted to the permissive temperature of 25°C. Samples were taken at the indicated time points and the migration shift of Mde4-13Myc (upper panel) and cell cycle progression (lower panel) were determined by western blot analysis and microscopy respectively. Lysates from wild-type cells (No tag), asynchronous mde4-13Myc (Asy), and mde4-13Myc cells arrested in mitosis using the nda3-KM311 mutation (nda3-KM311) are also shown. (D) Immunoprecipitated Mde4-13Myc from _mde4-13Myc clp1_Δ cells arrested in metaphase using the nda3-KM311 mutation was treated with buffer alone, recombinant MBP (Maltose binding protein), MBP-Clp1, or lambda phosphatase (λ PPase) then analyzed using western blotting. (E) In vitro kinase assays were performed using Cdk1 immunoprecipitated, using Cdc13 (S. pombe cyclin B) antibodies, from metaphase arrested nda3-KM311 cells, and bacterially expressed 6His-Mde4 as substrate. Protein labeled by γ-32P was detected with a Phospho Imager (Molecular Dynamics), and the gel was stained with Coomassie Blue (CB) as a loading control. The level of Cdk1 was determined by western blotting.
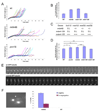
Figure 2. Clp1 promotes loading of the monopolin onto the spindle during anaphase
(A) mde4-GFP cells expressing mRFP-α-tubulin were imaged by fluorescence microscopy. (B) pcs1-GFP cdc11-GFP (upper panel) and _clp1_Δ pcs1-GFP cdc11-GFP (lower panel) cells were analyzed using fluorescent time-lapse microscopy. Images were collected at one minute intervals, beginning immediately prior to entry into mitosis. The SPB marker Cdc11-GFP was used to monitor cell cycle progression. At time zero both cells are in interphase just prior to mitotic entry. The arrow indicates anaphase onset, when Pcs1 begins to localize to the spindle. (C) Mde4-GFP localization is shown in wild-type (left panels) and _clp1_Δ cells (right panels). Interphase cells are shown in the top row, and cells from early to late anaphase are shown in the lower panels. Cells were grown asynchronously, fixed, stained with DAPI, and imaged by fluorescence microscopy.
Figure 3. Monopolin mutants have delays in anaphase onset and show occasional spindle collapse
(A) Spindle elongation was monitored in wild-type and monopolin mutant cells. Spindle length were measured every one minute from time-lapse sequences of asynchronous GFP-atb2 (n=17), _GFP-atb2 mde4_Δ (n=14), and _GFP-atb2 pcs1_Δ (n=8) cells. (B) Asynchronous wild-type and _mde4_Δ cells expressing Mad2-GFP were scored for the presence of Mad2-GFP puncta at kinetochores. Error bars represent standard deviation. (C) Genetic interactions between the mde4Δ, mde4-12A, and mde4-12D mutations and spindle assembly checkpoint mutations. The _mde4_Δ, mde4-12A and mde4-12D mutants were crossed with the spindle assembly checkpoint mutants _bub1_Δ, _mad1_Δ, _mad2_Δ and _mad3_Δ. Genetic interactions are shown as synthetic lethality (S.L), strong growth defect (+/−) and normal growth (+++). (D) A comparison of spindle lengths of wild-type and monopolin mutant cells at the metaphase/anaphase transition. The mean lengths of the spindle were compared between wild-type and _pcs1_Δ (single asterisks, p<0.01), _mde4_Δ (double asterisk, p<0.0001), or mde4-12A (triple asterisk p<0.0001), as well as _mde4_Δ and mde4-12A (quadruple asterisk p<0.0001) by a t test. Error bars represent standard deviation. (E) Time-lapse analysis using GFP-tubulin expressing wild-type (upper panel) and _mde4_Δ (lower panel) mutant cells is shown. The images were collected at one minute intervals. (F) An example of the chromosome co-segregation phenotype observed in _mde4_Δ cells is shown (left panel, arrow indicates septum position). The frequency of anaphase cells with lagging chromosomes and septated cells with co-segregated chromosomes is shown for wild-type and _mde4_Δ cells (right panel). Error bars represent standard deviation.
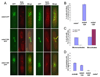
Figure 4. Mde4-12A prematurely localizes to the spindle instead of kinetochores before anaphase onset and displays lagging chromosomes in anaphase
(A) Localization of Mde4, Mde4-12A, and Mde4-12D at metaphase was examined in the following strains: mde4-GFP sid4-mRFP, mde4-12A-GFP sid4-mRFP, mde4-12D-GFP sid4mRFP (left panel), mde4-GFP nuf2-mRFP, mde4-12A-GFP nuf2-mRFP, and mde4-12D-GFP nuf2-mRFP (right panel). Cells were arrested in metaphase by overexpression of Mad2 using the pREP3X-Mad2 plasmid for 19 hr in the absence of thiamine. Cells were fixed and imaged by fluorescence microscopy. The yellow arrow indicates a kinetochore that does not appear to label with Mde4-12A-GFP. (B) The frequency of Mde4 localization to the metaphase spindle was determined. mde4-GFP sid4-mRFP, mde4-12A-GFP sid4-mRFP, mde4-12D-GFP sid4-mRFP cells were arrested at metaphase by Mad2 overexpression for 19h. Cells showing spindle localization were counted among metaphase arrested cells exhibiting unseparated condensed chromosomes and separated spindle pole bodies. Error bars represent standard deviation. (C) Quantification of Pcs1 spindle localization in pre-anaphase, and post-anaphase mde4+ and mde4-12A cells. pcs1-GFP and pcs1-GFP mde4-12A cells were grown asynchronously at 30°C then fixed and stained with DAPI. Of cells that showed Pcs1 spindle localization, the frequency that were mononucleate or binucleate is shown. Error bars represent standard deviation. (D) Frequency of lagging chromosomes in mde4 mutants was determined by counting the percent of anaphase cells with lagging or unevenly segregated chromosomes from asynchronously growing _mde4_Δ, mde4-12A, mde4-12D, and wild-type cells. Error bars represent standard deviation.
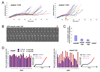
Figure 5. Mde4-12D localizes poorly to anaphase spindles
(A) Early and late anaphase localization of wild-type Mde4-GFP, Mde4-12A-GFP, Mde4-12D-GFP (upper panel), and Pcs1-GFP in mde4+, mde4-12A, and mde4-12D cells (lower panel) is shown. (B) Early (top row) and late anaphase (bottom row) localization of Mde4-12A-GFP in _clp1_Δ and Pcs1-GFP in _mde4-12A clp1_Δ cells is shown. In both panels (A) and (B), cells were grown asynchronously, fixed, and imaged by fluorescence microscopy.
Figure 6. Both mde4-12A and mde4-12D cells display distinct defects in spindle elongation
(A) Spindle elongation was monitored in mde4-12A and mde4-12D cells. Spindle length was measured every one minute from time-lapse sequences of asynchronous GFP-atb2 mde4-12A (n=20) and GFP-atb2 mde4-12D (n=17) cells. Arrow indicates spindle collapse in the cell shown in part B. (B) Spindle collapse and re-elongation was observed in 1 of 17 the GFP-atb2 mde-12D cells shown part A. Images were collected every minute and spindle collapse was reconfirmed by examining a 3D view of the spindle from stacked images (data not shown) at the time point of spindle shortening (red arrow). (C) Frequency of chromosome co-segregation in mde4 mutants was determined by counting the percent of septated cells with a single DNA mass in only one daughter cell. Error bars represent standard deviation. (D) Comparison of spindle elongation rate between mde4+ (n=15) and mde4-12D (n=14) cells. Spindle elongation rates were obtained by measuring the slope of plots of spindle length over time using linear regression in individual mde4+ and mde4-12D anaphase cells. Slopes were calculated for early (spindle lengths of 3.5um to 6um, shown as a blue bar) and late (spindle lengths of 6 to 10 um, shown as a red bar) anaphase. Each pair of red and blue bars represents an individual cell.
Figure 7. Model of phosphorylation-dependent regulation of chromosome segregation by the monopolin complex
Mde4-Pcs1 (green) localize to the nucleolus (green spot in nucleus) and the clustered kinetochores (smaller green spot at periphery of the nucleus) in interphase. In early mitosis Mde4 becomes phosphorylated (probably by Cdk1), which is required to maintain Pcs1-Mde4 at kinetochores (3 small green spots) to promote proper attachment of microtubules (MTs) to kinetochores to prevent lagging chromosomes in anaphase. In anaphase, Clp1 and other phosphatases dephosphorylate Mde4 to promote loading of monopolin onto the spindle (spindle shown in orange) to stabilize it and allow for proper elongation to prevent chromosome cosegregation.
Comment in
- Chromosome segregation: monopolin goes spindle.
Khmelinskii A, Schiebel E. Khmelinskii A, et al. Curr Biol. 2009 Jun 23;19(12):R482-4. doi: 10.1016/j.cub.2009.05.006. Curr Biol. 2009. PMID: 19549493
Similar articles
- Chromosome segregation: monopolin goes spindle.
Khmelinskii A, Schiebel E. Khmelinskii A, et al. Curr Biol. 2009 Jun 23;19(12):R482-4. doi: 10.1016/j.cub.2009.05.006. Curr Biol. 2009. PMID: 19549493 - Chromosome segregation: monopolin attracts condensin.
Dudas A, Polakova S, Gregan J. Dudas A, et al. Curr Biol. 2011 Aug 23;21(16):R634-6. doi: 10.1016/j.cub.2011.06.059. Curr Biol. 2011. PMID: 21855006 Free PMC article. - Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation.
Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E. Khmelinskii A, et al. Dev Cell. 2009 Aug;17(2):244-56. doi: 10.1016/j.devcel.2009.06.011. Dev Cell. 2009. PMID: 19686685 - Assembling the spindle midzone in the right place at the right time.
Khmelinskii A, Schiebel E. Khmelinskii A, et al. Cell Cycle. 2008 Feb 1;7(3):283-6. doi: 10.4161/cc.7.3.5349. Epub 2007 Nov 21. Cell Cycle. 2008. PMID: 18235228 Review. - Factors that Control Mitotic Spindle Dynamics.
Fraschini R. Fraschini R. Adv Exp Med Biol. 2017;925:89-101. doi: 10.1007/5584_2016_74. Adv Exp Med Biol. 2017. PMID: 27722958 Review.
Cited by
- The molecular basis of monopolin recruitment to the kinetochore.
Plowman R, Singh N, Tromer EC, Payan A, Duro E, Spanos C, Rappsilber J, Snel B, Kops GJPL, Corbett KD, Marston AL. Plowman R, et al. Chromosoma. 2019 Sep;128(3):331-354. doi: 10.1007/s00412-019-00700-0. Epub 2019 Apr 30. Chromosoma. 2019. PMID: 31037469 Free PMC article. - Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation.
Chen JS, Lu LX, Ohi MD, Creamer KM, English C, Partridge JF, Ohi R, Gould KL. Chen JS, et al. J Cell Biol. 2011 Nov 14;195(4):583-93. doi: 10.1083/jcb.201105074. Epub 2011 Nov 7. J Cell Biol. 2011. PMID: 22065639 Free PMC article. - Merotelic kinetochore attachment: causes and effects.
Gregan J, Polakova S, Zhang L, Tolić-Nørrelykke IM, Cimini D. Gregan J, et al. Trends Cell Biol. 2011 Jun;21(6):374-81. doi: 10.1016/j.tcb.2011.01.003. Epub 2011 Feb 8. Trends Cell Biol. 2011. PMID: 21306900 Free PMC article. Review. - Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast).
Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B. Carpy A, et al. Mol Cell Proteomics. 2014 Aug;13(8):1925-36. doi: 10.1074/mcp.M113.035824. Epub 2014 Apr 23. Mol Cell Proteomics. 2014. PMID: 24763107 Free PMC article. - Monopolin recruits condensin to organize centromere DNA and repetitive DNA sequences.
Burrack LS, Applen Clancey SE, Chacón JM, Gardner MK, Berman J. Burrack LS, et al. Mol Biol Cell. 2013 Sep;24(18):2807-19. doi: 10.1091/mbc.E13-05-0229. Epub 2013 Jul 24. Mol Biol Cell. 2013. PMID: 23885115 Free PMC article.
References
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. - PubMed
- Cimini D. Detection and correction of merotelic kinetochore orientation by Aurora B and its partners. Cell Cycle. 2007;6:1558–1564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM068786/GM/NIGMS NIH HHS/United States
- R01 GM068786-04/GM/NIGMS NIH HHS/United States
- R01 GM068786-05/GM/NIGMS NIH HHS/United States
- R01GM068786/GM/NIGMS NIH HHS/United States
- R01 GM068786-06/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous