Distinctive chromatin in human sperm packages genes for embryo development - PubMed (original) (raw)
. 2009 Jul 23;460(7254):473-8.
doi: 10.1038/nature08162. Epub 2009 Jun 14.
Affiliations
- PMID: 19525931
- PMCID: PMC2858064
- DOI: 10.1038/nature08162
Distinctive chromatin in human sperm packages genes for embryo development
Saher Sue Hammoud et al. Nature. 2009.
Abstract
Because nucleosomes are widely replaced by protamine in mature human sperm, the epigenetic contributions of sperm chromatin to embryo development have been considered highly limited. Here we show that the retained nucleosomes are significantly enriched at loci of developmental importance, including imprinted gene clusters, microRNA clusters, HOX gene clusters, and the promoters of stand-alone developmental transcription and signalling factors. Notably, histone modifications localize to particular developmental loci. Dimethylated lysine 4 on histone H3 (H3K4me2) is enriched at certain developmental promoters, whereas large blocks of H3K4me3 localize to a subset of developmental promoters, regions in HOX clusters, certain noncoding RNAs, and generally to paternally expressed imprinted loci, but not paternally repressed loci. Notably, trimethylated H3K27 (H3K27me3) is significantly enriched at developmental promoters that are repressed in early embryos, including many bivalent (H3K4me3/H3K27me3) promoters in embryonic stem cells. Furthermore, developmental promoters are generally DNA hypomethylated in sperm, but acquire methylation during differentiation. Taken together, epigenetic marking in sperm is extensive, and correlated with developmental regulators.
Figures
Figure 1
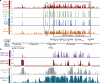
Developmental genes are associated with particular chromatin attributes in human sperm. GoMiner was used to identify enriched categories, and all categories displayed have an FDR < 0.01. The top five general categories are listed, after omitting nearly identical/redundant classes. An expanded GO table with the unfiltered top 30-60 categories, the total genes, number of changed genes, enrichment, and FDR are provided in the Supplemental Material.
Figure 2
Profiling of nucleosomes and their modifications at HOXD. For high-throughput sequencing, we display the mapped sequencing reads from D1 or a donor pool (red or orange bars; normalized difference score), and their significance (green or blue bars; FDR of 20 is <1% and FDR of 30 is <0.1%). a, The HOXD locus (black box) and an uncharacterized flanking locus (green oval). b, Profiling of nucleosome modifications at HOXD (in part a). The y-axis is signal intensity (log2, for ChIP-chip), or the normalized difference score for sequencing. The regions not tiled on the array are underlined in red.
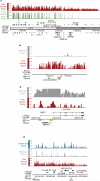
Figure 3
Nucleosomes enrichment at imprinted gene clusters, with high H3K4me3 at paternally-expressed non-coding RNAs, and paternally-demethylated regions. a, Histone enrichment at the 11p15.5 imprinted cluster (ending near OSBPL5), but not in the adjacent region. b and c, An expanded view of the DMRs (yellow rectangles) of H19 (paternally-methylated) and MEST (paternally-demethylated). d, High H3K4me3 at the promoters of the paternally-expressed genes BEGAIN, DLK1, and RTL, and the lack of H3K4me3 at the methylated intergenic-differentially methylated region (IG-DMR) of GTL2 in sperm. Notably, both H3K4me3 and H3K27me3 reside at the promoter of GTL2, which later acquires methylation in the embryo.
Figure 4
Developmental promoters in sperm lack DNA methylation, but acquire methylation during development. a, DNA methylation of the HOXD locus in the mature sperm (blue bars) or primary fibroblasts (orange line overlay). The y-axis is the signal intensity (log2) and the x-axis is the annotated physical map (HG17). The regions not tiled on the array are underlined in red.
Comment in
- Sperm histones are organized to carry epigenetic clues into the zygote.
Pfeifer GP, Szabo PE. Pfeifer GP, et al. Epigenomics. 2009 Oct;1(1):23. Epigenomics. 2009. PMID: 22423379 No abstract available.
Similar articles
- Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men.
Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. Hammoud SS, et al. Hum Reprod. 2011 Sep;26(9):2558-69. doi: 10.1093/humrep/der192. Epub 2011 Jun 17. Hum Reprod. 2011. PMID: 21685136 Free PMC article. - Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm.
Wu SF, Zhang H, Cairns BR. Wu SF, et al. Genome Res. 2011 Apr;21(4):578-89. doi: 10.1101/gr.113167.110. Epub 2011 Mar 7. Genome Res. 2011. PMID: 21383318 Free PMC article. - Chromatin alterations during the epididymal maturation of mouse sperm refine the paternally inherited epigenome.
Bedi YS, Roach AN, Thomas KN, Mehta NA, Golding MC. Bedi YS, et al. Epigenetics Chromatin. 2022 Jan 6;15(1):2. doi: 10.1186/s13072-021-00433-4. Epigenetics Chromatin. 2022. PMID: 34991687 Free PMC article. - Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics.
Miller D, Brinkworth M, Iles D. Miller D, et al. Reproduction. 2010 Feb;139(2):287-301. doi: 10.1530/REP-09-0281. Epub 2009 Sep 16. Reproduction. 2010. PMID: 19759174 Review. - Nucleosomes in mammalian sperm: conveying paternal epigenetic inheritance or subject to reprogramming between generations?
Gaspa-Toneu L, Peters AH. Gaspa-Toneu L, et al. Curr Opin Genet Dev. 2023 Apr;79:102034. doi: 10.1016/j.gde.2023.102034. Epub 2023 Mar 7. Curr Opin Genet Dev. 2023. PMID: 36893482 Free PMC article. Review.
Cited by
- Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells.
Luense LJ, Wang X, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, Berger SL. Luense LJ, et al. Epigenetics Chromatin. 2016 Jun 21;9:24. doi: 10.1186/s13072-016-0072-6. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27330565 Free PMC article. - Human sperm chromatin epigenetic potential: genomics, proteomics, and male infertility.
Castillo J, Estanyol JM, Ballescá JL, Oliva R. Castillo J, et al. Asian J Androl. 2015 Jul-Aug;17(4):601-9. doi: 10.4103/1008-682X.153302. Asian J Androl. 2015. PMID: 25926607 Free PMC article. Review. - PGC7, H3K9me2 and Tet3: regulators of DNA methylation in zygotes.
Kang J, Kalantry S, Rao A. Kang J, et al. Cell Res. 2013 Jan;23(1):6-9. doi: 10.1038/cr.2012.117. Epub 2012 Aug 7. Cell Res. 2013. PMID: 22868271 Free PMC article. - Sperm-inherited H3K27me3 epialleles are transmitted transgenerationally in cis.
Kaneshiro KR, Egelhofer TA, Rechtsteiner A, Cockrum C, Strome S. Kaneshiro KR, et al. Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2209471119. doi: 10.1073/pnas.2209471119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161922 Free PMC article. - Topology of chromosome centromeres in human sperm nuclei with high levels of DNA damage.
Wiland E, Fraczek M, Olszewska M, Kurpisz M. Wiland E, et al. Sci Rep. 2016 Aug 25;6:31614. doi: 10.1038/srep31614. Sci Rep. 2016. PMID: 27558650 Free PMC article.
References
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–74. - PubMed
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–7. - PubMed
- Balhorn R, Brewer L, Corzett M. DNA condensation by protamine and arginine-rich peptides: analysis of toroid stability using single DNA molecules. Mol Reprod Dev. 2000;56:230–4. - PubMed
- Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–6. - PubMed
- Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases