Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line - PubMed (original) (raw)
Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line
Nelson C Lau et al. Genome Res. 2009 Oct.
Abstract
Piwi proteins, a subclass of Argonaute-family proteins, carry approximately 24-30-nt Piwi-interacting RNAs (piRNAs) that mediate gonadal defense against transposable elements (TEs). We analyzed the Drosophila ovary somatic sheet (OSS) cell line and found that it expresses miRNAs, endogenous small interfering RNAs (endo-siRNAs), and piRNAs in abundance. In contrast to intact gonads, which contain mixtures of germline and somatic cell types that express different Piwi-class proteins, OSS cells are a homogenous somatic cell population that expresses only PIWI and primary piRNAs. Detailed examination of its TE-derived piRNAs and endo-siRNAs revealed aspects of TE defense that do not rely upon ping-pong amplification. In particular, we provide evidence that a subset of piRNA master clusters, including flamenco, are specifically expressed in OSS and ovarian follicle cells. These data indicate that the restriction of certain TEs in somatic gonadal cells is largely mediated by a primary piRNA pathway.
Figures
Figure 1.
Cation-exchange chromatography enriches for Argonaute- and Piwi-enclosed RNAs. (A) Phase contrast image of OSS cells illustrating their sheet-like morphology; under high density they also tend to form clumps. (B) pCp labeling of RNAs from total OSS RNA and various elutions from a HiTrap Q column. The mobility of the RNAs in the extract and elutions is slightly different from that of the synthetic markers owing to different salt concentrations of the loaded samples. Highly abundant 2S rRNA is visible in the input and ran slightly larger than its 30-nt sequence. In the flowthrough, ∼21–24-nt RNAs are visible, inferred to include miRNAs and siRNAs. In the 0.3 M salt elution, ∼24–28-nt RNAs are visible, inferred to represent the piRNA fraction. A heterogenous ladder of RNA fragments elutes with 1.0 M salt. Libraries were constructed from the flowthrough and 0.3 M salt elution.
Figure 2.
miRNA expression in OSS cells. (A) OSS cells predominantly express a characteristic population of miRNAs (left) that overlaps only partially with the most abundant miRNAs annotated from across Drosophila development using 454 Life Sciences (Roche) sequencing (Ruby et al. 2007a). However, analysis of all reads indicates that the 454-annotated miRNAs and OSS miRNAs are very highly overlapping (right). Therefore, numerous ostensibly tissue-specific Drosophila miRNAs were captured at low levels in OSS cells by deep sequencing. (B) Five novel miRNA loci, expressed at a level of more than five reads/14 million library reads, were identified in OSS cells. Mature products were highlighted in green, and star strands in red; for some hairpins, the two small RNA products were cloned nearly equivalently.
Figure 3.
TE-piRNAs and TE-siRNAs in OSS cells. (A) Reproducibility of overall read distribution in the four OSS sequencing reactions. Analysis of raw reads is shown; the normalized data are completely overlapping (Supplemental Fig. S1). (B) Combined OSS library data. Blue depicts the size distribution across all reads, orange depicts miRNA reads, and red depicts reads that mapped to transposons; the latter clearly segment into a siRNA population (peaking at 21 nt) and a piRNA population (∼24–30 nt). (C–G) Combined OSS read distribution mapped to various families of TEs (as annotated by RepeatMasker). (Red) Antisense reads; (blue) sense reads. Two graphs are shown for each TE family, depicting the size distribution of reads that map uniquely to the genome and the distribution of reads that map to the genome 10 or more times. Various patterns are observed across the TEs with respect to piRNAs vs. siRNAs, or the fraction of unique vs. multiply-matching reads. The complete analysis of all read bins across all TE families is available in Supplemental Figure S2.
Figure 4.
Sequence properties of TE-siRNAs and TE-piRNAs in OSS cells. (A–D) Aggregate nucleotide composition of all TE-derived reads of length 21 nt (siRNA) or 27 nt (as a representative piRNA size); in all graphs, the _x_-axis represents nucleotide position along the piRNA, while the _y_-axis represents the percent nucleotide composition and each position. 5′ U bias is observed for bulk AS-TE-siRNAs as well as S- and AS-TE-piRNAs, but is lacking in bulk S-TE-siRNAs. Note also that S-TE-piRNAs lack any bias for adenine at position 10 (above the adenine bias of neighboring nucleotide positions), as would be expected for ping-pong pairs. Analysis of other TE read sizes are available in Supplemental Figure S3. (E–H) Specific analysis of roo reads shows similar trends. Analysis of all other individual TEs are available in Supplemental Figure S4. (I) Overlap analysis of piRNAs with three hypothetical configurations of distance between 5′ ends of a piRNA and its nearest neighbor on the opposite strand; +10 offset is typical of piRNA ping-pong, while −16 offset is consistent with a phased arrangement of ping-pong pairs (gray arrows depict “missing” piRNAs). (J) Analysis of ovary (black) and 0–2-h embryo (blue) piRNAs reveals strong ping-pong (+10 offset pairs) and modest evidence for a phased ping-pong pairs. No ping-pong is observed for OSS piRNAs (red).
Figure 5.
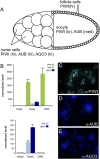
OSS cells are related to ovarian follicle cells. (A) Schematic of an individual mature germarium. The germline consists of 15 nurse cells and the developing oocyte; the latter is ensheathed by follicle cells of somatic origin. The accumulation of the three Piwi-class proteins—PIWI, AUB, and AGO3—in these celltypes is indicated. (B) Quantitative RT–PCR analysis of Piwi-class transcripts in OSS cells; ovaries were used as a positive control and female heads as a negative control. Transcript levels were normalized to RpL32 as a control, and expressed as the fold change above the level in heads. OSS cells express high levels of piwi, but not aub or AGO3. (C–E) Immunostaining of OSS cells with antibodies against the three Piwi-class proteins (green) verifies sole expression of PIWI; DNA was counterstained with DAPI (blue).
Figure 6.
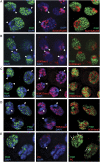
PIWI protein localization in OSS cells appears nucleoplasmic and not specific for any chromatin state. Triple staining of PIWI and DAPI with SU(VAR)205 (also known as HP1a) (A), H3K9me3 (B), H3K27me3 (C), H3K4me3 (D), and Polycomb (PC) (E). Arrowheads mark the location of the chromocenter, a DAPI-dense congregation of heterochromatin. PIWI is specifically absent from the chromocenter and does not overlap appreciably with either markers of silent or active chromatin. Some PIWI foci are near Polycomb foci, but these appear to be incidental and are never overlapping (arrows, E).
Similar articles
- Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the drosophila germline.
Shpiz S, Ryazansky S, Olovnikov I, Abramov Y, Kalmykova A. Shpiz S, et al. PLoS Genet. 2014 Feb 6;10(2):e1004138. doi: 10.1371/journal.pgen.1004138. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24516406 Free PMC article. - The capacity of target silencing by Drosophila PIWI and piRNAs.
Post C, Clark JP, Sytnikova YA, Chirn GW, Lau NC. Post C, et al. RNA. 2014 Dec;20(12):1977-86. doi: 10.1261/rna.046300.114. Epub 2014 Oct 21. RNA. 2014. PMID: 25336588 Free PMC article. - MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells.
Mugat B, Akkouche A, Serrano V, Armenise C, Li B, Brun C, Fulga TA, Van Vactor D, Pélisson A, Chambeyron S. Mugat B, et al. PLoS Genet. 2015 May 19;11(5):e1005194. doi: 10.1371/journal.pgen.1005194. eCollection 2015 May. PLoS Genet. 2015. PMID: 25993106 Free PMC article. - The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review. - piRNA pathway and the potential processing site, the nuage, in the Drosophila germline.
Pek JW, Patil VS, Kai T. Pek JW, et al. Dev Growth Differ. 2012 Jan;54(1):66-77. doi: 10.1111/j.1440-169x.2011.01316.x. Dev Growth Differ. 2012. PMID: 23741748 Review.
Cited by
- Divergent composition and transposon-silencing activity of small RNAs in mammalian oocytes.
Hou L, Liu W, Zhang H, Li R, Liu M, Shi H, Wu L. Hou L, et al. Genome Biol. 2024 Mar 26;25(1):80. doi: 10.1186/s13059-024-03214-w. Genome Biol. 2024. PMID: 38532500 Free PMC article. - Reactivation of a somatic errantivirus and germline invasion in Drosophila ovaries.
Yoth M, Maupetit-Méhouas S, Akkouche A, Gueguen N, Bertin B, Jensen S, Brasset E. Yoth M, et al. Nat Commun. 2023 Sep 29;14(1):6096. doi: 10.1038/s41467-023-41733-5. Nat Commun. 2023. PMID: 37773253 Free PMC article. - What Are the Functional Roles of Piwi Proteins and piRNAs in Insects?
Santos D, Feng M, Kolliopoulou A, Taning CNT, Sun J, Swevers L. Santos D, et al. Insects. 2023 Feb 14;14(2):187. doi: 10.3390/insects14020187. Insects. 2023. PMID: 36835756 Free PMC article. Review. - Non-coding RNAs in immunoregulation and autoimmunity: Technological advances and critical limitations.
Kumar D, Sahoo SS, Chauss D, Kazemian M, Afzali B. Kumar D, et al. J Autoimmun. 2023 Jan;134:102982. doi: 10.1016/j.jaut.2022.102982. Epub 2022 Dec 31. J Autoimmun. 2023. PMID: 36592512 Free PMC article. Review. - Resources and Methods for the Analysis of MicroRNA Function in Drosophila.
Mukherjee S, Sokol N. Mukherjee S, et al. Methods Mol Biol. 2022;2540:79-92. doi: 10.1007/978-1-0716-2541-5_3. Methods Mol Biol. 2022. PMID: 35980573 Review.
References
- Aravin A, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. - PubMed
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. - PubMed
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01-HG004261/HG/NHGRI NIH HHS/United States
- U01 HG004261/HG/NHGRI NIH HHS/United States
- K99-HD057298-01/HD/NICHD NIH HHS/United States
- R01 GM083300/GM/NIGMS NIH HHS/United States
- K99 HD057298/HD/NICHD NIH HHS/United States
- R01-GM083300/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials