Dereplication and de novo sequencing of nonribosomal peptides - PubMed (original) (raw)
Dereplication and de novo sequencing of nonribosomal peptides
Julio Ng et al. Nat Methods. 2009 Aug.
Abstract
Nonribosomal peptides (NRPs) are of great pharmacological importance, but there is currently no technology for high-throughput NRP 'dereplication' and sequencing. We used multistage mass spectrometry followed by spectral alignment algorithms for sequencing of cyclic NRPs. We also developed an algorithm for comparative NRP dereplication that establishes similarities between newly isolated and previously identified similar but nonidentical NRPs, substantially reducing dereplication efforts.
Figures
Figure 1
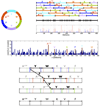
Experimental and theoretical spectra of seglitide Cyclic(N-methyl-Ala, Tyr, D-Trp, Lys, Val, Phe (a) Cyclic diagram of seglitide. A+14 denotes a methylated alanine; the integer residue masses are 85, 163, 186, 128, 99 and 147. (b) MS2 fragmentation of seglitide generates up to 6 linear peptides representing different linearized variants of the same cyclic peptide. (c) The theoretical spectrum for seglitide is a superposition of the fragment masses from the linearized peptide. (d) Experimental spectrum of seglitide (the peaks corresponding to prefix masses are shown in red). (e) The auto-convolution of the spectrum in insert d has prominent peaks for offsets corresponding to masses of amino acids (shown in red). The peak at 0 is truncated. (f) Generation of a gapped peptide from a theoretical spectrum of seglitide. The theoretical spectrum is colored to highlight various linear peptides. For illustration purposes only 3 linearized (A+14YWKV (blue), FA+14YWKV (red) and VFA+14YWK (green)) versions of the cyclic peptide are shown. The frequent 2-amino-acid tag YW is observed in 3 different locations in the spectrum. Additionally, the offsets between 3 consecutive locations of tag YW reveal the masses of amino acids F and V. (g) The gapped peptide constructed from f combines YW (derived from a frequent tag) with VF (derived from the inter distances between tag locations). A+14 and K are inferred from the flanking masses of YW and VF. The complete sequence A+14YWKVF is recovered, but gaps may be generated.
Similar articles
- Nonribosomal peptides: from genes to products.
Schwarzer D, Finking R, Marahiel MA. Schwarzer D, et al. Nat Prod Rep. 2003 Jun;20(3):275-87. doi: 10.1039/b111145k. Nat Prod Rep. 2003. PMID: 12828367 Review. - NRPquest: Coupling Mass Spectrometry and Genome Mining for Nonribosomal Peptide Discovery.
Mohimani H, Liu WT, Kersten RD, Moore BS, Dorrestein PC, Pevzner PA. Mohimani H, et al. J Nat Prod. 2014 Aug 22;77(8):1902-9. doi: 10.1021/np500370c. Epub 2014 Aug 12. J Nat Prod. 2014. PMID: 25116163 Free PMC article. - Application of an efficient gene targeting system linking secondary metabolites to their biosynthetic genes in Aspergillus terreus.
Guo CJ, Knox BP, Sanchez JF, Chiang YM, Bruno KS, Wang CC. Guo CJ, et al. Org Lett. 2013 Jul 19;15(14):3562-5. doi: 10.1021/ol401384v. Epub 2013 Jul 10. Org Lett. 2013. PMID: 23841722 Free PMC article. - Nonribosomal peptide synthesis in animals: the cyclodipeptide synthase of Nematostella.
Seguin J, Moutiez M, Li Y, Belin P, Lecoq A, Fonvielle M, Charbonnier JB, Pernodet JL, Gondry M. Seguin J, et al. Chem Biol. 2011 Nov 23;18(11):1362-8. doi: 10.1016/j.chembiol.2011.09.010. Chem Biol. 2011. PMID: 22118670 - [Advances in the study of the mechanism and application of nonribosomal peptide synthetases].
Wang SY. Wang SY. Wei Sheng Wu Xue Bao. 2007 Aug;47(4):734-7. Wei Sheng Wu Xue Bao. 2007. PMID: 17944384 Review. Chinese.
Cited by
- Discovery and biosynthesis of cyclic plant peptides via autocatalytic cyclases.
Chigumba DN, Mydy LS, de Waal F, Li W, Shafiq K, Wotring JW, Mohamed OG, Mladenovic T, Tripathi A, Sexton JZ, Kautsar S, Medema MH, Kersten RD. Chigumba DN, et al. Nat Chem Biol. 2022 Jan;18(1):18-28. doi: 10.1038/s41589-021-00892-6. Epub 2021 Nov 22. Nat Chem Biol. 2022. PMID: 34811516 - NPS: scoring and evaluating the statistical significance of peptidic natural product-spectrum matches.
Tagirdzhanov AM, Shlemov A, Gurevich A. Tagirdzhanov AM, et al. Bioinformatics. 2019 Jul 15;35(14):i315-i323. doi: 10.1093/bioinformatics/btz374. Bioinformatics. 2019. PMID: 31510666 Free PMC article. - Toward a global picture of bacterial secondary metabolism.
Seyedsayamdost MR. Seyedsayamdost MR. J Ind Microbiol Biotechnol. 2019 Mar;46(3-4):301-311. doi: 10.1007/s10295-019-02136-y. Epub 2019 Jan 25. J Ind Microbiol Biotechnol. 2019. PMID: 30684124 Free PMC article. - Mining for Microbial Gems: Integrating Proteomics in the Postgenomic Natural Product Discovery Pipeline.
Du C, van Wezel GP. Du C, et al. Proteomics. 2018 Sep;18(18):e1700332. doi: 10.1002/pmic.201700332. Epub 2018 Jun 10. Proteomics. 2018. PMID: 29708658 Free PMC article. Review. - Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra.
Gurevich A, Mikheenko A, Shlemov A, Korobeynikov A, Mohimani H, Pevzner PA. Gurevich A, et al. Nat Microbiol. 2018 Mar;3(3):319-327. doi: 10.1038/s41564-017-0094-2. Epub 2018 Jan 22. Nat Microbiol. 2018. PMID: 29358742 Free PMC article.
References
- Sieber SA, Marahiel MA. Molecular Mechanisms Underlying Nonribosomal Peptide Synthesis: Approaches to New Antibiotics. Chem. Rev. 2005;105:715–738. - PubMed
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. Journal of Natural Products. 2007;70:461–477. - PubMed
- Hamada T, Matsunaga S, Yano G, Fusetani N. Polytheonamides A and B, Highly Cytotoxic, Linear Polypeptides with Unprecedented Structural Features, from the Marine Sponge, Theonella swinhoei. J. Am. Chem. Soc. 2005;127:110–118. - PubMed
- Ireland CM, Durso AR, Newman RA, Hacker MP. Antineoplastic Cyclic Peptides from the Marine Tunicate Lissoclinum patella. J. Org. Chem. 1982;47:360–361.
- Li J, Burgett A, Esser L, Amezcua C, Harran P. Total synthesis of nominal diazonamides: Part 2. on the true structure and origin of natural isolates. Angew. Chem Intl. Ed. Engl. 2001;40:4770–4773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 1-P41-RR024851-01/RR/NCRR NIH HHS/United States
- R01 GM086283/GM/NIGMS NIH HHS/United States
- P41 GM103484/GM/NIGMS NIH HHS/United States
- R01 GM086283-01/GM/NIGMS NIH HHS/United States
- U19 CA052955/CA/NCI NIH HHS/United States
- P41 RR024851/RR/NCRR NIH HHS/United States
- GM086283/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous