The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP - PubMed (original) (raw)
The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP
Qingfei Jiang et al. Nature. 2009.
Abstract
DNA-damage-induced SOS mutations arise when Escherichia coli DNA polymerase (pol) V, activated by a RecA nucleoprotein filament (RecA*), catalyses translesion DNA synthesis. Here we address two longstanding enigmatic aspects of SOS mutagenesis, the molecular composition of mutagenically active pol V and the role of RecA*. We show that RecA* transfers a single RecA-ATP stoichiometrically from its DNA 3'-end to free pol V (UmuD'(2)C) to form an active mutasome (pol V Mut) with the composition UmuD'(2)C-RecA-ATP. Pol V Mut catalyses TLS in the absence of RecA* and deactivates rapidly upon dissociation from DNA. Deactivation occurs more slowly in the absence of DNA synthesis, while retaining RecA-ATP in the complex. Reactivation of pol V Mut is triggered by replacement of RecA-ATP from RecA*. Thus, the principal role of RecA* in SOS mutagenesis is to transfer RecA-ATP to pol V, and thus generate active mutasomal complex for translesion synthesis.
Figures
Figure 1. DNA synthesis by pol V Mut or pol V transactivated by RecA*
The p/t DNA is a hairpin containing a 3-nucleotide template overhang. a, b, DNA synthesis by pol V Mut–(RecA E38KΔC17) (a) or pol V Mut–(wild-type RecA) (b) in the absence of RecA*. c, d, DNA synthesis by pol V transactivated by RecA E38KΔC17* (c) or wild-type RecA* (d). e, f, DNA synthesis by pol V Mut–(RecA E38KΔC17) (e) or pol V Mut–(wild-type RecA) (f) undergoing deactivation as a function of time. g, Deactivation of pol V Mut–(RecA E38KΔC17) and pol V Mut–(wild-type RecA) measured by quantifying DNA synthesis obtained from data in panels e and f, respectively. Complete reactivation of deactivated pol V Mut is observed by addition of trans RecA* at each delay time. h, pol V Mut performs one round of DNA synthesis and cannot reinitiate synthesis on separate p/t DNA substrate. The concentration of p/t DNA is 1 μM. We estimate that the fraction of active polymerase in the reaction is 50% for pol V Mut–(RecA E38KΔC17) and 25% for pol V Mut–(wild-type RecA). i, Addition of trans RecA E38KΔC17* enables pol V Mut–(RecA E38KΔC17) to reinitiate DNA synthesis on a separate p/t DNA substrate. Cycling also occurs using pol V Mut–(wild-type RecA) in conjunction with trans wild-type RecA* (data not shown). Trans RecA* is at 1 μM when present.
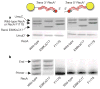
Figure 2. Activated pol V Mut complex is composed of UmuD′2C–RecA–ATP
a, b, DNA synthesis by pol V Mut–(wild-type RecA) (b), but not pol V Mut–(RecA E38KΔC17) (a), requires the presence of ATPγS or ATP. c, d, Activated pol V Mut–(wild-type RecA) complex contains UmuC, UmuD′ and wild-type RecA, in a 1:1:1 ratio. e, Fl-wild-type RecA and ATPγS remain bound in deactivated pol V Mut–(Fl-wild-type RecA) complexes as measured by affinity chromatography. f, Reactivation of pol V Mut–(Fl-wild-type RecA) is achieved by replacing Fl-wild-type RecA and [35S]ATPγS bound to deactivated pol V Mut–(Fl-wild-type RecA) with unlabelled wild-type RecA and ATPγS from wild-type RecA*. Reactivated pol V Mut–(Fl-wild-type RecA) complexes were isolated as in panel e.
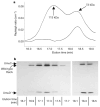
Figure 3. Determination of the molecular mass of pol V Mut–(wild-type RecA) by MALS
a, After wild-type-RecA*-mediated transactivation of UmuD′2C and removal of wild-type RecA*, the mixture of pol V Mut–(wild-type RecA) and non-activated pol V was resolved by size-exclusion chromatography (upper trace), and the molecular mass corresponding to each peak was measured by MALS. Non-activated pol V run separately on the silica gel elutes at 18.4 min (lower trace). b, Silver-stained SDS–polyacrylamide gel showing the protein composition from the two peaks contained in panel a (upper trace).
Figure 4. Pol V activation requires transfer of a RecA molecule from the DNA 3′ end of RecA*
a, The amount of RecA transferred to pol V from exposed 3′- or 5′-tips of RecA* are similar for wild-type RecA*, RecA E38KΔC17* and RecA F117S*. b, DNA synthesis catalysed by pol V Mut–(wild-type RecA) or pol V Mut–(RecA E38KΔC17) occurs when RecA is transferred from the 3′-RecA* tip (left gel). Pol V Mut cannot synthesize DNA when RecA transfer takes place from the 5′-RecA* tip (right gel). Mutagenically inactive RecA F117S* fails to activate pol V.
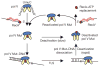
Figure 5. Model depicting pol V activation, deactivation and reactivation
Pol V (UmuD′2C) is barely active in the absence of RecA*. A molecule of RecA (red circle) and ATP (green triangle) is transferred from the DNA 3′-end of RecA* to form a mutasomal complex (pol V Mut) containing UmuD′2C–RecA–ATP; this catalyses TLS in the absence of RecA*. After TLS, pol V Mut undergoes rapid deactivation upon dissociation from DNA, with RecA and ATP retained in the complex. When pol V Mut is activated, we surmise that RecA–ATP is bound to UmuD′2, and that dissociation from p/t DNA triggers a repositioning of RecA–ATP to bind with UmuC to deactivate pol V Mut. Reactivation of pol V Mut involves a replacement of RecA–ATP from the RecA* 3′-tip. Free pol V Mut undergoes slow deactivation, with RecA and ATP retained in the complex. A damaged DNA base is shown as a black cross in the template strand.
Similar articles
- Conformational regulation of Escherichia coli DNA polymerase V by RecA and ATP.
Jaszczur MM, Vo DD, Stanciauskas R, Bertram JG, Sikand A, Cox MM, Woodgate R, Mak CH, Pinaud F, Goodman MF. Jaszczur MM, et al. PLoS Genet. 2019 Feb 4;15(2):e1007956. doi: 10.1371/journal.pgen.1007956. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30716079 Free PMC article. - DNA polymerase V and RecA protein, a minimal mutasome.
Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. Schlacher K, et al. Mol Cell. 2005 Feb 18;17(4):561-72. doi: 10.1016/j.molcel.2005.01.006. Mol Cell. 2005. PMID: 15721259 - Visualization of two binding sites for the Escherichia coli UmuD'(2)C complex (DNA pol V) on RecA-ssDNA filaments.
Frank EG, Cheng N, Do CC, Cerritelli ME, Bruck I, Goodman MF, Egelman EH, Woodgate R, Steven AC. Frank EG, et al. J Mol Biol. 2000 Mar 31;297(3):585-97. doi: 10.1006/jmbi.2000.3591. J Mol Biol. 2000. PMID: 10731413 - A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V.
Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. Patel M, et al. Crit Rev Biochem Mol Biol. 2010 Jun;45(3):171-84. doi: 10.3109/10409238.2010.480968. Crit Rev Biochem Mol Biol. 2010. PMID: 20441441 Free PMC article. Review. - Mutations for Worse or Better: Low-Fidelity DNA Synthesis by SOS DNA Polymerase V Is a Tightly Regulated Double-Edged Sword.
Jaszczur M, Bertram JG, Robinson A, van Oijen AM, Woodgate R, Cox MM, Goodman MF. Jaszczur M, et al. Biochemistry. 2016 Apr 26;55(16):2309-18. doi: 10.1021/acs.biochem.6b00117. Epub 2016 Apr 12. Biochemistry. 2016. PMID: 27043933 Free PMC article. Review.
Cited by
- RecA proteins from Deinococcus geothermalis and Deinococcus murrayi--cloning, purification and biochemical characterisation.
Wanarska M, Krawczyk B, Hildebrandt P, Kur J. Wanarska M, et al. BMC Mol Biol. 2011 Apr 22;12:17. doi: 10.1186/1471-2199-12-17. BMC Mol Biol. 2011. PMID: 21513512 Free PMC article. - New insights into the structures and interactions of bacterial Y-family DNA polymerases.
Timinskas K, Venclovas Č. Timinskas K, et al. Nucleic Acids Res. 2019 May 21;47(9):4393-4405. doi: 10.1093/nar/gkz198. Nucleic Acids Res. 2019. PMID: 30916324 Free PMC article. - ImuA Facilitates SOS Mutagenesis by Inhibiting RecA-Mediated Activity in Myxococcus xanthus.
Sheng D, Wang Y, Jiang Z, Liu D, Li Y. Sheng D, et al. Appl Environ Microbiol. 2021 Aug 26;87(18):e0091921. doi: 10.1128/AEM.00919-21. Epub 2021 Aug 26. Appl Environ Microbiol. 2021. PMID: 34190612 Free PMC article. - RecA-independent recombination: Dependence on the Escherichia coli RarA protein.
Jain K, Wood EA, Romero ZJ, Cox MM. Jain K, et al. Mol Microbiol. 2021 Jun;115(6):1122-1137. doi: 10.1111/mmi.14655. Epub 2020 Dec 19. Mol Microbiol. 2021. PMID: 33247976 Free PMC article. - Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination.
Kim T, Chitteni-Pattu S, Cox BL, Wood EA, Sandler SJ, Cox MM. Kim T, et al. PLoS Genet. 2015 Jun 5;11(6):e1005278. doi: 10.1371/journal.pgen.1005278. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26047498 Free PMC article.
References
- Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. - PubMed
- Friedberg EC, et al. DNA Repair and Mutagenesis. 2nd. ASM Press; 2006. pp. 463–555.
- Pham P, Bertram JG, O'Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. - PubMed
- Becherel OJ, Fuchs RP, Wagner J. Pivotal role of the β-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair. 2002;1:703–708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 GM032335/GM/NIGMS NIH HHS/United States
- R37 GM021422-33/GM/NIGMS NIH HHS/United States
- R37GM21422/GM/NIGMS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 ES012259/ES/NIEHS NIH HHS/United States
- R37 GM021422/GM/NIGMS NIH HHS/United States
- GM32335/GM/NIGMS NIH HHS/United States
- ES12259/ES/NIEHS NIH HHS/United States
- R01 GM032335/GM/NIGMS NIH HHS/United States
- R01 ES012259-20/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases