Recent progress in the biology and physiology of sirtuins - PubMed (original) (raw)
Review
Recent progress in the biology and physiology of sirtuins
Toren Finkel et al. Nature. 2009.
Abstract
The sirtuins are a highly conserved family of NAD(+)-dependent enzymes that regulate lifespan in lower organisms. Recently, the mammalian sirtuins have been connected to an ever widening circle of activities that encompass cellular stress resistance, genomic stability, tumorigenesis and energy metabolism. Here we review the recent progress in sirtuin biology, the role these proteins have in various age-related diseases and the tantalizing notion that the activity of this family of enzymes somehow regulates how long we live.
Figures
Figure 1. Mouse knockout models as tools for exploring sirtuin function
Gene targeting of SIRT1, SIRT3, SIRT4, SIRT5, SIRT6 and SIRT7 has been reported. The phenotypes include a reduction in median lifespan, ranging from a usual survival of days (SIRT1) to weeks (SIRT6) or months (SIRT7). In contrast, although biochemical phenotypes have been reported, _Sirt3_−/− and _Sirt4_−/− mice appear outwardly normal. Initial reports suggest that _Sirt5_−/− mice also exhibit no obvious phenotype.
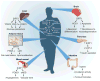
Figure 2. The diverse physiological roles of the sirtuins
Shown are examples of the organ-specific physiology of SIRT1, along with some of the direct or indirect targets of sirtuin regulation (see text for details). In addition, examples of some of the SIRT1-regulated intracellular parameters are presented, ranging from modulation of progenitor differentiation to altering the threshold for apoptosis. At the beginning of a compound name or process (for example, ‘Tumour formation’ at bottom right), down-arrow indicates ‘decreasing’, up-arrow indicates ‘increasing’ and ‘Δ’ indicates ‘change in’.
Figure 3. Complex regulation of SIRT1 activity
The promoter of SIRT1 is positively and negatively regulated by the binding of various transcription factors and repressors, including H1C1, CtBP, p53, FOXO3A and E2F1. The acetylation and hence activity of many of these factors are in turn controlled by SIRT1. The SIRT1 message is also regulated by the RNA binding protein HuR and the p53-regulated microRNA miR-34A. Finally, SIRT protein activity is regulated positively and negatively by interacting proteins such as AROS and DBC1, as well as by the overall metabolic state, as reflected in the NAD/NADH ratio.
Similar articles
- Sirtuins, epigenetics and longevity.
Wątroba M, Dudek I, Skoda M, Stangret A, Rzodkiewicz P, Szukiewicz D. Wątroba M, et al. Ageing Res Rev. 2017 Nov;40:11-19. doi: 10.1016/j.arr.2017.08.001. Epub 2017 Aug 5. Ageing Res Rev. 2017. PMID: 28789901 Review. - Sirtuins: a family of proteins with implications for human performance and exercise physiology.
Lappalainen Z. Lappalainen Z. Res Sports Med. 2011 Jan;19(1):53-65. doi: 10.1080/15438627.2011.536068. Res Sports Med. 2011. PMID: 21253976 Review. - The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity.
Satoh A, Stein L, Imai S. Satoh A, et al. Handb Exp Pharmacol. 2011;206:125-62. doi: 10.1007/978-3-642-21631-2_7. Handb Exp Pharmacol. 2011. PMID: 21879449 Free PMC article. Review. - Aging and cancer: are sirtuins the link?
Rodriguez RM, Fraga MF. Rodriguez RM, et al. Future Oncol. 2010 Jun;6(6):905-15. doi: 10.2217/fon.10.57. Future Oncol. 2010. PMID: 20528229 Review. - Sirtuins and their relevance to the kidney.
Hao CM, Haase VH. Hao CM, et al. J Am Soc Nephrol. 2010 Oct;21(10):1620-7. doi: 10.1681/ASN.2010010046. Epub 2010 Jul 1. J Am Soc Nephrol. 2010. PMID: 20595677 Free PMC article. Review.
Cited by
- Finding a sirtuin truth in Huntington's disease.
La Spada AR. La Spada AR. Nat Med. 2012 Jan 6;18(1):24-6. doi: 10.1038/nm.2624. Nat Med. 2012. PMID: 22227661 No abstract available. - The NAD-dependent deacetylase SIRT2 is required for programmed necrosis.
Narayan N, Lee IH, Borenstein R, Sun J, Wong R, Tong G, Fergusson MM, Liu J, Rovira II, Cheng HL, Wang G, Gucek M, Lombard D, Alt FW, Sack MN, Murphy E, Cao L, Finkel T. Narayan N, et al. Nature. 2012 Dec 13;492(7428):199-204. doi: 10.1038/nature11700. Epub 2012 Nov 28. Nature. 2012. PMID: 23201684 Retracted. - Mitochondrial metabolism, sirtuins, and aging.
Sack MN, Finkel T. Sack MN, et al. Cold Spring Harb Perspect Biol. 2012 Dec 1;4(12):a013102. doi: 10.1101/cshperspect.a013102. Cold Spring Harb Perspect Biol. 2012. PMID: 23209156 Free PMC article. Review. - Sirtuins, metabolism, and cancer.
Martinez-Pastor B, Mostoslavsky R. Martinez-Pastor B, et al. Front Pharmacol. 2012 Feb 21;3:22. doi: 10.3389/fphar.2012.00022. eCollection 2012. Front Pharmacol. 2012. PMID: 22363287 Free PMC article. - The SIRT1/TP53 axis is activated upon B-cell receptor triggering via miR-132 up-regulation in chronic lymphocytic leukemia cells.
Dal Bo M, D'Agaro T, Gobessi S, Zucchetto A, Dereani S, Rossi D, Zaja F, Pozzato G, Di Raimondo F, Gaidano G, Laurenti L, Del Poeta G, Efremov DG, Gattei V, Bomben R. Dal Bo M, et al. Oncotarget. 2015 Aug 7;6(22):19102-17. doi: 10.18632/oncotarget.3905. Oncotarget. 2015. PMID: 26036258 Free PMC article.
References
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S cerevisiae. Cell. 1995;80:485–496. - PubMed
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. - PubMed
- Guarente L, Picard F. Calorie restriction—the SIR2 connection. Cell. 2005;120:473–482. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources