Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations - PubMed (original) (raw)
Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations
Hannah Carter et al. Cancer Res. 2009.
Abstract
Large-scale sequencing of cancer genomes has uncovered thousands of DNA alterations, but the functional relevance of the majority of these mutations to tumorigenesis is unknown. We have developed a computational method, called Cancer-specific High-throughput Annotation of Somatic Mutations (CHASM), to identify and prioritize those missense mutations most likely to generate functional changes that enhance tumor cell proliferation. The method has high sensitivity and specificity when discriminating between known driver missense mutations and randomly generated missense mutations (area under receiver operating characteristic curve, >0.91; area under Precision-Recall curve, >0.79). CHASM substantially outperformed previously described missense mutation function prediction methods at discriminating known oncogenic mutations in P53 and the tyrosine kinase epidermal growth factor receptor. We applied the method to 607 missense mutations found in a recent glioblastoma multiforme sequencing study. Based on a model that assumed the glioblastoma multiforme mutations are a mixture of drivers and passengers, we estimate that 8% of these mutations are drivers, causally contributing to tumorigenesis.
Figures
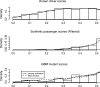
Figure 1. Principal components analysis of nsSNPs vs. synthetic passenger mutations
Synthetic passenger mutations (red) and high MAF nsSNPs from the HapMap project (blue) have substantial overlap in the space defined by principal components one, three, and four, but there are regions in the space occupied only by high MAF nsSNPs and regions occupied only by synthetic passengers.
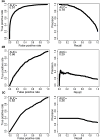
Figure 2. ROC and PR curves calculated for A) CHASM, B) PolyPhen PSIC, and C) SIFT on the training set mutations
CHASM training out-of-bag scores were used to generate the ROC and PR curves in A). A color version is available as Supplementary Figure 6.
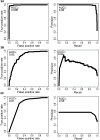
Figure 3. ROC and PR curves calculated for A) CHASM, B) PolyPhen PSIC, and C) SIFT on TP53 and synthetic passenger mutations held out of the CHASM training set
A color version is available as Supplementary Figure 7.
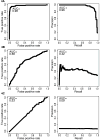
Figure 4. ROC and PR curves calculated for A) CHASM, B) PolyPhen PSIC, and C) SIFT on EGFR and synthetic passenger mutations held out of the CHASM training set
A color version is available as Supplementary Figure 8.
Figure 5. Histograms of CHASM scores for driver mutations and passenger mutations held out from the training set, and 607 mutations experimentally identified in GBM
Estimated kernel density for each set of scores (solid line) and fitted mixture of the driver and passenger score densities (dashed line) are shown superimposed on the histograms.
Similar articles
- CanDrA: cancer-specific driver missense mutation annotation with optimized features.
Mao Y, Chen H, Liang H, Meric-Bernstam F, Mills GB, Chen K. Mao Y, et al. PLoS One. 2013 Oct 30;8(10):e77945. doi: 10.1371/journal.pone.0077945. eCollection 2013. PLoS One. 2013. PMID: 24205039 Free PMC article. - Predicting the functional consequences of somatic missense mutations found in tumors.
Carter H, Karchin R. Carter H, et al. Methods Mol Biol. 2014;1101:135-59. doi: 10.1007/978-1-62703-721-1_8. Methods Mol Biol. 2014. PMID: 24233781 - Assessment of computational methods for predicting the effects of missense mutations in human cancers.
Gnad F, Baucom A, Mukhyala K, Manning G, Zhang Z. Gnad F, et al. BMC Genomics. 2013;14 Suppl 3(Suppl 3):S7. doi: 10.1186/1471-2164-14-S3-S7. Epub 2013 May 28. BMC Genomics. 2013. PMID: 23819521 Free PMC article. - Computational Approaches to Prioritize Cancer Driver Missense Mutations.
Zhao F, Zheng L, Goncearenco A, Panchenko AR, Li M. Zhao F, et al. Int J Mol Sci. 2018 Jul 20;19(7):2113. doi: 10.3390/ijms19072113. Int J Mol Sci. 2018. PMID: 30037003 Free PMC article. Review. - Looking beyond drivers and passengers in cancer genome sequencing data.
De S, Ganesan S. De S, et al. Ann Oncol. 2017 May 1;28(5):938-945. doi: 10.1093/annonc/mdw677. Ann Oncol. 2017. PMID: 27998972 Review.
Cited by
- Predicting the functional consequences of cancer-associated amino acid substitutions.
Shihab HA, Gough J, Cooper DN, Day IN, Gaunt TR. Shihab HA, et al. Bioinformatics. 2013 Jun 15;29(12):1504-10. doi: 10.1093/bioinformatics/btt182. Epub 2013 Apr 25. Bioinformatics. 2013. PMID: 23620363 Free PMC article. - Utilizing protein structure to identify non-random somatic mutations.
Ryslik GA, Cheng Y, Cheung KH, Modis Y, Zhao H. Ryslik GA, et al. BMC Bioinformatics. 2013 Jun 13;14:190. doi: 10.1186/1471-2105-14-190. BMC Bioinformatics. 2013. PMID: 23758891 Free PMC article. - CDMPred: a tool for predicting cancer driver missense mutations with high-quality passenger mutations.
Wang L, Sun H, Yue Z, Xia J, Li X. Wang L, et al. PeerJ. 2024 Sep 6;12:e17991. doi: 10.7717/peerj.17991. eCollection 2024. PeerJ. 2024. PMID: 39253604 Free PMC article. - Predicting cancer-associated germline variations in proteins.
Martelli PL, Fariselli P, Balzani E, Casadio R. Martelli PL, et al. BMC Genomics. 2012 Jun 18;13 Suppl 4(Suppl 4):S8. doi: 10.1186/1471-2164-13-S4-S8. BMC Genomics. 2012. PMID: 22759656 Free PMC article. - Dipeptide analysis of p53 mutations and evolution of p53 family proteins.
Huang Q, Yu L, Levine AJ, Nussinov R, Ma B. Huang Q, et al. Biochim Biophys Acta. 2014 Jan;1844(1 Pt B):198-206. doi: 10.1016/j.bbapap.2013.04.002. Epub 2013 Apr 10. Biochim Biophys Acta. 2014. PMID: 23583620 Free PMC article.
References
- Kaminker JS, Zhang Y, Waugh A, et al. Distinguishing Cancer-Associated Missense Mutations from Common Polymorphisms. Cancer Res. 2007;67(2):465–73. - PubMed
- Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–74. - PubMed
Publication types
MeSH terms
Grants and funding
- CA135877/CA/NCI NIH HHS/United States
- 28XS268/PHS HHS/United States
- CA62924/CA/NCI NIH HHS/United States
- CA43460/CA/NCI NIH HHS/United States
- R37 CA057345/CA/NCI NIH HHS/United States
- R21 CA135877-01/CA/NCI NIH HHS/United States
- R01 CA057345/CA/NCI NIH HHS/United States
- CA57345/CA/NCI NIH HHS/United States
- R37 CA043460/CA/NCI NIH HHS/United States
- CA121113/CA/NCI NIH HHS/United States
- P50 CA062924/CA/NCI NIH HHS/United States
- R21 CA135877/CA/NCI NIH HHS/United States
- R01 CA121113/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous