Co-transcriptional splicing of constitutive and alternative exons - PubMed (original) (raw)
Co-transcriptional splicing of constitutive and alternative exons
Amy Pandya-Jones et al. RNA. 2009 Oct.
Abstract
In metazoan organisms, pre-mRNA splicing is thought to occur during transcription, and it is postulated that these two processes are functionally coupled via still-unknown mechanisms. Current evidence supports co-transcriptional spliceosomal assembly, but there is little quantitative information on how much splicing is completed during RNA synthesis. Here we isolate nascent chromatin-associated RNA from free, nucleoplasmic RNA already released from the DNA template. Using a quantitative RT-PCR assay, we show that the majority of introns separating constitutive exons are already excised from the human c-Src and fibronectin pre-mRNAs that are still in the process of synthesis, and that these introns are removed in a general 5'-to-3' order. Introns flanking alternative exons in these transcripts are also removed during synthesis, but show differences in excision efficiency between cell lines with different regulatory conditions. Our data suggest that skipping of an exon can induce a lag in splicing compared to intron removal under conditions of exon inclusion. Nevertheless, excision of the long intron encompassing the skipped exon is still completed prior to transcript release into the nucleoplasm. Thus, we demonstrate that the decision to include or skip an alternative exon is made during transcription and not post-transcriptionally.
Figures
FIGURE 1.
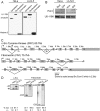
Purification of chromatin-associated pre-mRNA. (A) Chemiluminescent immunoblot of fractionated HeLa and LA-N-5 cell lines. Anti-α-tubulin, anti-U170K, and anti-Histone3 antibodies detect the cytoplasmic, nucleoplasmic, and chromatin fractions, respectively. Twenty-five micrograms of total protein from each fraction was loaded per lane. (B) Fluorescent immunoblot of HeLa and LA-N-5 chromatin and nucleoplasmic fractions. Twenty-five micrograms of total protein from each fraction was loaded per lane. An asterisk denotes hyperphosphorylated form of Pol II. (C) Diagrams of the c-Src and fibronectin gene structure. The 18 nt c-Src N1 exon is alternatively spliced and the 1A promoter is used in both HeLa and LA-N-5 cells. The fibronectin transcript contains exons 25 (EDII or EIIIB) and 33 (EDI or EIIIA), which are alternative. (D) Radiolabeled RT-PCR of cytoplasmic RNA showing inclusion levels of the c-Src N1, FN Exon 25 and 33 alternative exons in HeLa and LA-N-5 cell lines.
FIGURE 2.
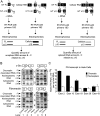
Exon abundance decreases with increasing proximity to 3′ end of the transcript within the chromatin fraction. (A) Depiction of quantitative RT-PCR protocol. HeLa and LA-N-5 chromatin-associated, nucleoplasmic, or cytoplasmic RNA was subjected to reverse transcription using a gene specific primer in the presence of 10−18 mol of in vitro transcribed (IVT) RNA internal controls. The amount of each analyzed exon–intron or exon–exon junction PCR product was calculated by comparison of band intensities between control and experimental samples. The fraction of each intron remaining in the three fractions was calculated by dividing the amount of the 5′ exon–intron junction product for each intron (the PCR product of primers 1 and 2) by the total product across the region (the sum of the intron product from primers 1 and 2 and the spliced product from primers 1 and 3). (B) Radiolabeled RT-PCR sample results for c-Src exons 10 and 11 and FN exon 35 and 36. Lanes 1,4 are control IVT RNA's as are the lower bands in lanes 2,3,5,6. Upper bands in lanes 2,5 (HeLa) and 3,6 (LA-N-5) represent the quantity of endogenous exon–intron or exon–exon junctions in each fraction of RNA. Asterisks denote nonspecific products. (C) Histogram representing the absolute amount of each analyzed constitutive FN exon in 1 μg of chromatin-associated or nucleoplasmic HeLa RNA. Error bars represent the standard error of mean (SEM).
FIGURE 3.
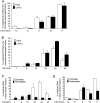
Introns flanking constitutive exons in c-Src and FN are excised co-transcriptionally. (A) Histogram representing the percentage of unspliced RNA across c-Src introns 1a, 1c, 8, 10, and 11 in chromatin-associated pre-mRNAs from HeLa (white) and LA-N-5 (black) cells. (B) Histogram representing the percentage of unspliced RNA across FN introns 1, 9, 18, 35, 42, and 45 in chromatin-associated pre-mRNA from HeLa (white) and LA-N-5 (black) cell lines. (C) Histogram representing the percentage of unspliced RNA across FN introns 1, 9, 18, 35, 42, and 45 per microgram of HeLa chromatin-associated (white) and nucleoplasmic (black) RNA. (D) Same as (C) except data represent introns in the c-Src transcript from chromatin-associated and nucleoplasmic fractions. Error bars represent the standard error of mean (SEM).
FIGURE 4.
The introns downstream from FN alternative exons 25 and 33 are excised prior to the upstream ones. (A) Analysis of FN exon 25 splicing intermediates. The two possible splicing intermediates generated during alternative exon inclusion are diagrammed on the left. Denaturing PAGE analysis of RT-PCR splicing intermediate products is pictured on the right. The lower bands in all gels, as well as the upper bands in lanes 1,4,7,10,13,16 are internal IVT controls. Upper bands in lanes 2,3,11,12 represent splicing intermediate products from HeLa and LA-N-5 chromatin-associated RNA. Lanes 5,6,14,15 and lanes 8,9,17,18 represent the same information from the nucleoplasmic and cytoplasmic RNA fractions, respectively. Control reactions are shown in Supplemental Figure S5. (B) Quantification of lanes 2,3,11,12 in A. (C) Analysis of FN exon 33 splicing intermediates. The experimental setup is identical to A, except primers target exon 33. (D) Quantification of lanes 2,3,11,12 in C. Error bars represent the standard error of mean (SEM).
FIGURE 5.
The intron upstream of c-Src exon N1 is excised prior to the downstream one. (A) Analysis of c-Src N1 splicing intermediates. The two possible splicing intermediates are diagrammed on the left. Denaturing PAGE analysis of RT-PCR splicing intermediate products is pictured on the right. The lower bands in all gels, as well as the upper bands in lanes 1,4,7,10,13,16 are internal controls. An asterisk denotes a nonspecific product. Upper bands in lanes 2,3,11,12 represent splicing intermediate products from HeLa and LA-N-5 chromatin-associated RNA. Lanes 5,6,14,15 and lanes 8,9,17,18 represent the same information except from the nucleoplasmic and cytoplasmic RNA fractions, respectively. Control reactions are shown in Supplemental Figure S5. (B) Quantification of lanes 2,3,11,12 in A. Asterisk indicates nonspecific product. (C) Stacked column graph showing percentage of the detectable splicing intermediate product found in LA-N-5 chromatin-associated and nucleoplasmic fractions normalized to the total amount of FN or c-Src RNA in each fraction. Error bars represent the standard error of mean (SEM).
FIGURE 6.
Splicing across c-Src exon N1 is inefficient when N1 is skipped. (A) Histogram displaying the percentage of unspliced RNA across analyzed introns along the c-Src transcript in the HeLa chromatin-associated RNA fraction. Lanes 1,2,7–9 are the constitutive regions shown in Figure 3A. Lane 3 represents the percentage of unspliced RNA across the 5′ exon-intron junction of the intron upstream of N1 relative to the total spliced and unspliced RNA across exons 3–4. Lanes 4–6 indicate the amount of unspliced RNA across the other three exon-intron junctions. (B) Diagram of exon–intron junctions analyzed in A and method of quantification. (C) Same as in A, except analysis was done using LA-N-5 chromatin-associated RNA. (D) Same as in B, except analysis accounts for the situation where N1 is included. (E) RT-PCR analysis of spliced exon 3-N1-4 products in HeLa and LA-N-5 chromatin-associated RNA fraction indicating that spliced RNA is present in HeLa chromatin-associated fractions. Error bars represent the standard error of mean (SEM).
FIGURE 7.
Splicing is efficient across the FN exon 25 and exon 33 alternative regions when the exons are completely included. (A) Histogram displaying the percentage of unspliced RNA across analyzed introns along the FN transcript in the HeLa chromatin-associated RNA fraction. Lanes 1–3,12–14 are the constitutive regions shown in Figure 3B. Lane 4 represents the percentage of unspliced RNA across the 5′ exon–intron junction of the intron upstream of exon 25 relative to the total amount RNA across exons 24–26. Lanes 5–7 indicate the amount of unspliced RNA across the other three exon–intron junctions. Similarly, lane 8 represents the percentage of unspliced RNA across the 5′ exon–intron junction of the intron upstream of exon 33 relative to the total amount RNA across exons 32–34. Lanes 9–11 indicate the amount of unspliced RNA across the other three exon–intron junctions. (B) Diagram of exon–intron junctions analyzed in A and C and method of quantification. (C) Same as in A, except analysis was done using LA-N-5 chromatin-associated RNA. (D) RT-PCR analysis of spliced exon 24–26 and 32–34 products in HeLa and LA-N-5 chromatin-associated RNA fraction. Error bars represent the standard error of mean (SEM).
Similar articles
- Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription.
Baurén G, Wieslander L. Baurén G, et al. Cell. 1994 Jan 14;76(1):183-92. doi: 10.1016/0092-8674(94)90182-1. Cell. 1994. PMID: 8287477 - A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon.
Modafferi EF, Black DL. Modafferi EF, et al. Mol Cell Biol. 1997 Nov;17(11):6537-45. doi: 10.1128/MCB.17.11.6537. Mol Cell Biol. 1997. PMID: 9343417 Free PMC article. - Exon and intron definition in pre-mRNA splicing.
De Conti L, Baralle M, Buratti E. De Conti L, et al. Wiley Interdiscip Rev RNA. 2013 Jan-Feb;4(1):49-60. doi: 10.1002/wrna.1140. Epub 2012 Oct 8. Wiley Interdiscip Rev RNA. 2013. PMID: 23044818 Review. - Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing.
Shkreta L, Bell B, Revil T, Venables JP, Prinos P, Elela SA, Chabot B. Shkreta L, et al. Cancer Treat Res. 2013;158:41-94. doi: 10.1007/978-3-642-31659-3_3. Cancer Treat Res. 2013. PMID: 24222354 Review.
Cited by
- Deciphering the plant splicing code: experimental and computational approaches for predicting alternative splicing and splicing regulatory elements.
Reddy AS, Rogers MF, Richardson DN, Hamilton M, Ben-Hur A. Reddy AS, et al. Front Plant Sci. 2012 Feb 7;3:18. doi: 10.3389/fpls.2012.00018. eCollection 2012. Front Plant Sci. 2012. PMID: 22645572 Free PMC article. - SplicePie: a novel analytical approach for the detection of alternative, non-sequential and recursive splicing.
Pulyakhina I, Gazzoli I, 't Hoen PA, Verwey N, den Dunnen JT, Aartsma-Rus A, Laros JF. Pulyakhina I, et al. Nucleic Acids Res. 2015 Jul 13;43(12):e80. doi: 10.1093/nar/gkv242. Epub 2015 Mar 23. Nucleic Acids Res. 2015. PMID: 25800735 Free PMC article. - Alternative splicing: a pivotal step between eukaryotic transcription and translation.
Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ. Kornblihtt AR, et al. Nat Rev Mol Cell Biol. 2013 Mar;14(3):153-65. doi: 10.1038/nrm3525. Epub 2013 Feb 6. Nat Rev Mol Cell Biol. 2013. PMID: 23385723 Review. - Splicing Regulation: A Molecular Device to Enhance Cancer Cell Adaptation.
Pagliarini V, Naro C, Sette C. Pagliarini V, et al. Biomed Res Int. 2015;2015:543067. doi: 10.1155/2015/543067. Epub 2015 Jul 26. Biomed Res Int. 2015. PMID: 26273627 Free PMC article. Review.
References
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. - PubMed
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - PubMed
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. - PubMed
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 1988;2:754–765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous