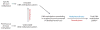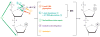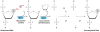Active DNA demethylation mediated by DNA glycosylases - PubMed (original) (raw)
Review
Active DNA demethylation mediated by DNA glycosylases
Jian-Kang Zhu. Annu Rev Genet. 2009.
Abstract
Active DNA demethylation is involved in many vital developmental and physiological processes of plants and animals. Recent genetic and biochemical studies in Arabidopsis have demonstrated that a subfamily of DNA glycosylases function to promote DNA demethylation through a base excision-repair pathway. These specialized bifunctional DNA glycosylases remove the 5-methylcytosine base and then cleave the DNA backbone at the abasic site, resulting in a gap that is then filled with an unmethylated cytosine nucleotide by as yet unknown DNA polymerase and ligase enzymes. Evidence suggests that active DNA demethylation in mammalian cells is also mediated at least in part by a base excision repair pathway where the AID/Apobec family of deaminases convert 5-methylcytosine to thymine followed by G/T mismatch repair by the DNA glycosylase MBD4 or TDG. This review also discusses other possible mechanisms of active DNA demethylation, how genome DNA methylation status might be sensed to regulate the expression of demethylase genes, and the targeting of demethylases by small RNAs.
Figures
Figure 1
Role of active DNA demethylation in establishing DNA methylation patterns. DNA methylation patterns are established by the combined actions of DNA methyltransferases and demethylases. Demethylases are required for pruning unwanted DNA methylation generated by promiscuous methyltransferases, and DNA methylation reprogramming/remodeling during development and in response to environmental changes.
Figure 2
Diagram showing various possible levels and mechanisms of active DNA demethylation. (a) Base excision repair (BER) initiated by 5-methylcytosine (5-meC) DNA glycosylase. This is the predominant mechanism in plants but may also function in mammals. (b) Base excision repair initiated by coupled activities of 5-meC deaminase that converts 5-meC to T, and G/T mismatch DNA glycosylase that corrects the G/T mismatch. This appears to be the predominant mechanism in mammals but may also play a role in plants. (c) Nucleotide excision repair that removes methylated CpG dinucleotides. (d ) Oxidative removal of the methyl group. (e) Hydrolytic removal of the methyl group, releasing it as methanol.
Figure 3
The Arabidopsis DNA demethylase ROS1 is required for preventing transcriptional silencing of genes that are under dynamic control by DNA methylation and demethylation. (a) Dynamic control of DNA methylation level and transcription activity by DNA methyltransferase and demethylase enzymes. (b) Silencing of the RD29A-LUC transgene in the ros1-1 mutant. Left, luminescence image of the ros1-1 mutant and WT plants; right, bright field illumination of all plants. The color scale under the luminescence image shows the luminescence intensity from black (lowest) to white (highest). Silencing in the ros1 mutant can be released by mutations in any component of the RNA-directed DNA methylation pathway, supporting dynamic control of the RD29A-LUC transgene by the opposing methylation and demethylation pathways. WT, wild type Arabidopsis seedlings.
Figure 4
Diagram of ROS1-mediated DNA demethylation by a base excision repair pathway. Question marks indicate as yet unidentified enzymes in the pathway. ROS1 is a bifunctional DNA glycosylase/lyase that removes the 5-methylcytosine base and then cleaves the DNA backbone at the abasic site, resulting in a gap that is then filled with an unmethylated cytosine nucleotide by as yet unknown DNA polymerase and ligase enzymes.
Similar articles
- Regulation and function of DNA methylation in plants and animals.
He XJ, Chen T, Zhu JK. He XJ, et al. Cell Res. 2011 Mar;21(3):442-65. doi: 10.1038/cr.2011.23. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321601 Free PMC article. Review. - Active DNA demethylation in plants and animals.
Zhang H, Zhu JK. Zhang H, et al. Cold Spring Harb Symp Quant Biol. 2012;77:161-73. doi: 10.1101/sqb.2012.77.014936. Epub 2012 Nov 28. Cold Spring Harb Symp Quant Biol. 2012. PMID: 23197304 Free PMC article. Review. - The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation.
Martínez-Macías MI, Córdoba-Cañero D, Ariza RR, Roldán-Arjona T. Martínez-Macías MI, et al. J Biol Chem. 2013 Feb 22;288(8):5496-505. doi: 10.1074/jbc.M112.427617. Epub 2013 Jan 11. J Biol Chem. 2013. PMID: 23316050 Free PMC article. - Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation.
Schomacher L, Han D, Musheev MU, Arab K, Kienhöfer S, von Seggern A, Niehrs C. Schomacher L, et al. Nat Struct Mol Biol. 2016 Feb;23(2):116-124. doi: 10.1038/nsmb.3151. Epub 2016 Jan 11. Nat Struct Mol Biol. 2016. PMID: 26751644 Free PMC article. - DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases.
Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. Morales-Ruiz T, et al. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6853-8. doi: 10.1073/pnas.0601109103. Epub 2006 Apr 19. Proc Natl Acad Sci U S A. 2006. PMID: 16624880 Free PMC article.
Cited by
- Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review. - Genome-Wide Analysis of DNA Demethylases in Land Plants and Their Expression Pattern in Rice.
Mao S, Xiao J, Zhao Y, Hou J, Li L. Mao S, et al. Plants (Basel). 2024 Jul 26;13(15):2068. doi: 10.3390/plants13152068. Plants (Basel). 2024. PMID: 39124186 Free PMC article. - Recent Advances in Studies of Genomic DNA Methylation and Its Involvement in Regulating Drought Stress Response in Crops.
Fan Y, Sun C, Yan K, Li P, Hein I, Gilroy EM, Kear P, Bi Z, Yao P, Liu Z, Liu Y, Bai J. Fan Y, et al. Plants (Basel). 2024 May 17;13(10):1400. doi: 10.3390/plants13101400. Plants (Basel). 2024. PMID: 38794470 Free PMC article. Review. - Transgenerational increases in DNA methylation in Arabidopsis plants defective in active DNA demethylation.
Tang K, Zhu X, Xie S, Lang Z, Zhu JK. Tang K, et al. Proc Natl Acad Sci U S A. 2024 May 28;121(22):e2320468121. doi: 10.1073/pnas.2320468121. Epub 2024 May 20. Proc Natl Acad Sci U S A. 2024. PMID: 38768356 Free PMC article. - Systematic Analysis of DNA Demethylase Gene Families in Foxtail Millet (Setaria italica L.) and Their Expression Variations after Abiotic Stresses.
Sun Y, Wang X, Di Y, Li J, Li K, Wei H, Zhang F, Su Z. Sun Y, et al. Int J Mol Sci. 2024 Apr 18;25(8):4464. doi: 10.3390/ijms25084464. Int J Mol Sci. 2024. PMID: 38674049 Free PMC article.
References
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–801. Provided evidence that the Arabidopsis ROS1 protein can specifically recognize and process 5-methylcytosine DNA through a DNA glycosylase mechanism. - PMC - PubMed
- Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. - PubMed
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–75. - PubMed
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–63. - PubMed
- Bender J. DNA methylation and epigenetics. Annu Rev Plant Biol. 2004;55:41–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous