Real-time in vivo imaging of p16Ink4a reveals cross talk with p53 - PubMed (original) (raw)
. 2009 Aug 10;186(3):393-407.
doi: 10.1083/jcb.200904105.
Akiko Takahashi, Fumiko Hirota, Rika Nakayama, Naozumi Ishimaru, Yoshiaki Kubo, David J Mann, Masako Ohmura, Atsushi Hirao, Hideyuki Saya, Seiji Arase, Yoshio Hayashi, Kazuki Nakao, Mitsuru Matsumoto, Naoko Ohtani, Eiji Hara
Affiliations
- PMID: 19667129
- PMCID: PMC2728398
- DOI: 10.1083/jcb.200904105
Real-time in vivo imaging of p16Ink4a reveals cross talk with p53
Kimi Yamakoshi et al. J Cell Biol. 2009.
Abstract
Expression of the p16(Ink4a) tumor suppressor gene, a sensor of oncogenic stress, is up-regulated by a variety of potentially oncogenic stimuli in cultured primary cells. However, because p16(Ink4a) expression is also induced by tissue culture stress, physiological mechanisms regulating p16(Ink4a) expression remain unclear. To eliminate any potential problems arising from tissue culture-imposed stress, we used bioluminescence imaging for noninvasive and real-time analysis of p16(Ink4a) expression under various physiological conditions in living mice. In this study, we show that oncogenic insults such as ras activation provoke epigenetic derepression of p16(Ink4a) expression through reduction of DNMT1 (DNA methyl transferase 1) levels as a DNA damage response in vivo. This pathway is accelerated in the absence of p53, indicating that p53 normally holds the p16(Ink4a) response in check. These results unveil a backup tumor suppressor role for p16(Ink4a) in the event of p53 inactivation, expanding our understanding of how p16(Ink4a) expression is regulated in vivo.
Figures
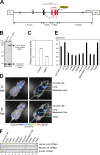
Figure 1.
Generation of human p16Ink4a reporter mice. (A) A large genomic DNA segment (195.4 kb) of human chromosome that contains the entire INK4a/ARF gene locus and surrounding sequences, including a putative DNA replication origin (RD) known to regulate p16Ink4a gene expression (Gonzalez et al., 2006), was engineered to express luciferase-tagged p16Ink4a. (B) BAC vector containing p16-luc DNA or empty BAC vector was transfected into 293T cells. Expression of the p16Ink4a-luciferase fusion protein was analyzed by Western blotting after selection with antibiotics. (C) BAC vector containing p16-luc DNA was introduced into 293T cells with or without BMI-1 expression plasmid along with 0.2 mg of MMLV (Moloney murine leukaemia virus)-lacZ plasmid. Luciferase activities were normalized by lacZ activities. Error bars indicate SD. (D) The 6-wk-old p16-luc mice were subjected to noninvasive BLI. Representative images of five different experiments are shown (left). The same mice were incised through the mouth and anus under anesthesia. Representative BLI data of five different experiments are shown. The color bar indicates photons with minimum and maximum threshold values. (E) Bioluminescence intensity emitted from the organs was graphed (log10 scale). The mean ± SD of five independent experiments is shown. (F) The levels of exogenous (human) p16Ink4a gene expression and endogenous (mouse) p16Ink4a gene expression in p16-luc mice were analyzed by semiquantitative RT-PCR. β-Actin was used as a loading control. Representative data of five different experiments are shown.
Figure 2.
Induction of human p16Ink4a gene expression during the onset of cellular senescence in cultured MEFs. (A and B) Primary MEFs derived from the p16-luc mice were rendered senescent by either serial passage (A) or oncogenic Ras expression (retrovirus infection; B). The levels of endogenous p16Ink4a expression and luciferase activity were measured. Representative data of three independent experiments are shown. As confirmation of senescence, representative photographs of the cells stained for SA β-gal activity are shown. β-Actin was used as a loading control. The means ± SD of three independent experiments are shown. WB, Western blot.
Figure 3.
The dynamics of p16Ink4a gene expression during the aging process in vivo_._ (A) The same p16-luc mice were subjected to noninvasive BLI every 2 wk throughout their entire life span. Representative images of 10 independent mice are shown. (B) The p16-luc mice (young and old) were subjected to noninvasive BLI. The same mice were then incised through mouth and anus under anesthesia. (A and B) The color bars indicate photons with minimum and maximum threshold values. (C) Bioluminescence intensity emitted from the organs from young (1.5 mo) and old (22.5 mo) mice were graphed (log10 scale). The mean ± SD of five independent experiments is shown. (D) The levels of exogenous (human) p16Ink4a gene expression and endogenous (mouse) p16Ink4a gene expression from young (Y; 1.5 mo) and old (O; 22.5 mo) mice were analyzed by semiquantitative RT-PCR. β-Actin was used as a loading control. Representative data of five independent experiments are shown.
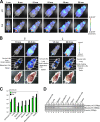
Figure 4.
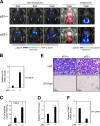
The dynamics of p16Ink4a gene expression during skin papilloma formation. (A) The p16-luc mice treated with DMBA/TPA were subjected to noninvasive BLI at the indicated time points after initiation of TPA treatment. Representative images of 10 independent experiments are shown (top). These papillomas were photographed in dimmed light (bottom). The color bar indicates photons with minimum and maximum threshold values. (B) Representative Western blots of biopsy samples of skin papillomas or control normal skin are shown using the antibodies indicated on the left. Vinculin was used as a loading control. (C) Hematoxylin and eosin (HE) staining, SA β-gal staining, and immunohistochemistry for endogenous p16Ink4a expression, phosphorylation of pRb at Ser807, Ki67 expression, phosphorylation of histone H2AX (γ-H2AX), and phosphorylation of p53 at Ser18 were performed by using biopsy samples of skin papillomas or control normal skin. The boxes denote regions shown below at higher magnification (p16 staining). (D) The levels of ROS were measured by using biopsy samples of skin papillomas or control normal skin. The means ± SD of three independent experiments are shown.
Figure 5.
Bisulfite sequence analysis of the mouse p16Ink4a gene promoter. A schematic illustration of the mouse p16Ink4a gene promoter is shown at the top, with the sequence numbered backwards from immediately before the initiating Met codon (the positions of the numbers are indicated by vertical dashes). The transcriptional start site is indicated by a thick vertical line, with the direction of transcription shown by the associated arrow. The coding region of the mouse p16Ink4a gene is annotated and indicated with a closed box. The clustering of CpG dinucleotides is shown with thin vertical lines through the promoter region. The positions of the primers for methyl-specific PCR are indicated below with two small arrows. Bisulfite-treated DNA was prepared from normal skin or early or late papillomas (Fig. 4). Methylation-specific PCR products were subcloned into the pGEM-T vector, and 10 clones for each sample were sequenced. Representative results of three different experiments are shown.
Figure 6.
Correlation between DNMT1 level and H3K9me2 level around the p16Ink4a gene promoter. (A) Early passage TIG-3 cells were infected with retrovirus encoding oncogenic Ras (+H-RasV12). Cells were then subjected to Western blot (WB) analysis at the indicated times with the antibodies shown on the left and to analysis of intracellular levels of ROS. (B) Early passage TIG-3 cells were infected with retrovirus encoding oncogenic Ras (RasV12) or control empty vector for 10 d and were subjected to Western blotting with the antibodies shown, analysis of intracellular levels of ROS, and to ChIP analysis using the antibodies indicated (IP). The precipitated DNA was amplified by real-time quantitative PCR (qPCR) using primers specific for the p16Ink4a gene promoter described previously (Bracken et al., 2007). (C and D) Early passage TIG-3 cells were infected with retrovirus encoding shRNA against DNMT1 or control scramble shRNA. Cell extracts were prepared from cells at 7 d after selection with puromycin and were subjected to Western blotting and quantitative real-time RT-PCR (RT-qPCR) analysis for p16Ink4a gene expression (C) and subjected to ChIP analysis using antibodies against H3K9me2 (D). (D) The precipitated DNA was amplified by real-time quantitative PCR using primers specific for the p16Ink4a gene promoter described previously (Bracken et al., 2007). (A–D) The means ± SD of three independent experiments are shown.
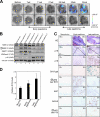
Figure 7.
Accelerated induction of p16Ink4a gene expression by DNA damage in mice lacking p53. (A) 8-wk-old p16-luc mice lacking the p53 gene (p53−/−) or their wild-type controls (p53+/+) were injected i.p. with 20 µg/g DXR and subjected to noninvasive BLI at the indicated time points. The same mice were then sacrificed, tissues were rapidly removed and placed in culture dishes, and ex vivo tissue BLI was performed. The tissues examined were as follows: 1, brain; 2, cervical LNs; 3, heart; 4, lung; 5, thymus; 6, small intestine; 7, liver; 8, kidney; 9, spleen. The color bars indicate photons with minimum and maximum threshold values. Representative images of five different experiments are shown. Schematic drawings illustrating the mouse bodies are shown (dotted lines). (B and C) Isolated tissues were subjected to analysis of quantitative real-time RT-PCR (RT-qPCR) for p16Ink4a gene expression (B) or to immunofluorescence analysis using antibody against γ-H2AX (red; C). DNA was stained with DAPI (blue). The histogram indicates the percentage of nuclei that were positive for γ-H2AX staining. The means ± SD of three independent experiments are shown. (C) Enlarged images of the boxed areas are shown below. Small intestinal crypts are marked by dashed lines. (D) Early passage TIG-3 cells were infected with retrovirus encoding shRNA against p53 or control scramble shRNA. Cell extracts were prepared after selection with puromycin and subjected to Western blotting using the antibodies shown on the left. β-Actin was used as a loading control.
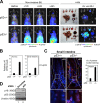
Figure 8.
Induction of p16Ink4a gene expression in p53 knockout mice. (A) p16-luc mice lacking the p53 gene (p53−/−) or their wild-type controls (p53+/+) were subjected to noninvasive BLI at the indicated age. The same mice were then incised under anesthesia (photographed under regular light) and subjected to BLI again (invasive BLI). Representative images of five different experiments are shown. The color bars indicate photons with minimum and maximum threshold values. Schematic drawings illustrating the mouse bodies are shown (dotted lines). The arrows show bioluminescence signals derived from thymus. (B–F) Thymus tissue was isolated from both genotypes of 10-wk-old mice (A) and subjected to analysis of quantitative real-time RT-PCR (RT-qPCR) for p16Ink4a gene expression (B) or DNMT1 gene expression (F) or histochemistry (hematoxylin and eosin [H&E] staining and SA β-gal staining; E), γ-H2AX foci (C), or intracellular levels of ROS (D). The means ± SD of three independent experiments are shown.
Figure 9.
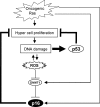
Cross talk between the p53 and p16 pathways through DDR. Although oncogenic Ras signaling has a potential to activate p16Ink4a gene expression, this effect is initially counteracted by an elevation of DNMT1 level and thereby causes a strong proliferative burst, resulting in the accumulation of DNA damage. The accumulation of DNA damage activates ROS production, which in turn blocks DNMT1 gene expression, thereby causing epigenetic derepression of p16Ink4a gene expression and thus senescence cell cycle arrest. This pathway is counterbalanced by the p53 pathway because p53 is immediately activated by DNA damage and blocks proliferation of damaged cells that cause further accumulation of DNA damage. Thus, the DDR pathway activating p16Ink4a expression is accelerated in the event of p53 inactivation.
Similar articles
- p16INK4A enhances the transcriptional and the apoptotic functions of p53 through DNA-dependent interaction.
Al-Khalaf HH, Nallar SC, Kalvakolanu DV, Aboussekhra A. Al-Khalaf HH, et al. Mol Carcinog. 2017 Jul;56(7):1687-1702. doi: 10.1002/mc.22627. Epub 2017 Mar 6. Mol Carcinog. 2017. PMID: 28218424 Free PMC article. - Resistance of primary cultured mouse hepatic tumor cells to cellular senescence despite expression of p16(Ink4a), p19(Arf), p53, and p21(Waf1/Cip1).
Obata M, Imamura E, Yoshida Y, Goto J, Kishibe K, Yasuda A, Ogawa K. Obata M, et al. Mol Carcinog. 2001 Sep;32(1):9-18. doi: 10.1002/mc.1059. Mol Carcinog. 2001. PMID: 11568971 - P16(INK4A) is implicated in both the immediate and adaptative response of human keratinocytes to UVB irradiation.
Chazal M, Marionnet C, Michel L, Mollier K, Dazard JE, Della Valle V, Larsen CJ, Gras MP, Basset-Séguin N. Chazal M, et al. Oncogene. 2002 Apr 18;21(17):2652-61. doi: 10.1038/sj.onc.1205349. Oncogene. 2002. PMID: 11965538 - p16(Ink4a) and senescence-associated β-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli.
Hall BM, Balan V, Gleiberman AS, Strom E, Krasnov P, Virtuoso LP, Rydkina E, Vujcic S, Balan K, Gitlin II, Leonova KI, Consiglio CR, Gollnick SO, Chernova OB, Gudkov AV. Hall BM, et al. Aging (Albany NY). 2017 Aug 2;9(8):1867-1884. doi: 10.18632/aging.101268. Aging (Albany NY). 2017. PMID: 28768895 Free PMC article. - Recovery of function in osteoarthritic chondrocytes induced by p16INK4a-specific siRNA in vitro.
Zhou HW, Lou SQ, Zhang K. Zhou HW, et al. Rheumatology (Oxford). 2004 May;43(5):555-68. doi: 10.1093/rheumatology/keh127. Epub 2004 Mar 16. Rheumatology (Oxford). 2004. PMID: 15026580
Cited by
- Manipulating senescence in health and disease: emerging tools.
Cheng S, Rodier F. Cheng S, et al. Cell Cycle. 2015;14(11):1613-4. doi: 10.1080/15384101.2015.1039359. Cell Cycle. 2015. PMID: 25928330 Free PMC article. No abstract available. - Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model.
Burd CE, Sorrentino JA, Clark KS, Darr DB, Krishnamurthy J, Deal AM, Bardeesy N, Castrillon DH, Beach DH, Sharpless NE. Burd CE, et al. Cell. 2013 Jan 17;152(1-2):340-51. doi: 10.1016/j.cell.2012.12.010. Cell. 2013. PMID: 23332765 Free PMC article. - Separate and combined effects of advanced age and obesity on mammary adipose inflammation, immunosuppression and tumor progression in mouse models of triple negative breast cancer.
Smith LA, Craven DM, Rainey MA, Cozzo AJ, Carson MS, Glenny EM, Sheth N, McDonell SB, Rezeli ET, Montgomery SA, Bowers LW, Coleman MF, Hursting SD. Smith LA, et al. Front Oncol. 2023 Jan 4;12:1031174. doi: 10.3389/fonc.2022.1031174. eCollection 2022. Front Oncol. 2023. PMID: 36686775 Free PMC article. - RNaseH2A downregulation drives inflammatory gene expression via genomic DNA fragmentation in senescent and cancer cells.
Sugawara S, Okada R, Loo TM, Tanaka H, Miyata K, Chiba M, Kawasaki H, Katoh K, Kaji S, Maezawa Y, Yokote K, Nakayama M, Oshima M, Nagao K, Obuse C, Nagayama S, Takubo K, Nakanishi A, Kanemaki MT, Hara E, Takahashi A. Sugawara S, et al. Commun Biol. 2022 Dec 28;5(1):1420. doi: 10.1038/s42003-022-04369-7. Commun Biol. 2022. PMID: 36577784 Free PMC article. - DNA Damage Regulates Senescence-Associated Extracellular Vesicle Release via the Ceramide Pathway to Prevent Excessive Inflammatory Responses.
Hitomi K, Okada R, Loo TM, Miyata K, Nakamura AJ, Takahashi A. Hitomi K, et al. Int J Mol Sci. 2020 May 25;21(10):3720. doi: 10.3390/ijms21103720. Int J Mol Sci. 2020. PMID: 32466233 Free PMC article.
References
- Bachman K.E., Park B.H., Rhee I., Rajagopalan H., Herman J.G., Baylin S.B., Kinzler K.W., Vogelstein B. 2003. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene.Cancer Cell. 3:89–95 - PubMed
- Ballester M., Castelló A., Ibáñez E., Sánchez A., Folch J.M. 2004. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals.Biotechniques. 37:610–613 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous