Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast - PubMed (original) (raw)
. 2009 Sep;11(9):1116-20.
doi: 10.1038/ncb1925. Epub 2009 Aug 16.
Affiliations
- PMID: 19684576
- PMCID: PMC2752306
- DOI: 10.1038/ncb1925
Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast
Soni Lacefield et al. Nat Cell Biol. 2009 Sep.
Abstract
Accurate chromosome segregation depends on the kinetochore, which is the complex of proteins that link microtubules to centromeric DNA. The kinetochore of the budding yeast Saccharomyces cerevisiae consists of more than 80 proteins assembled on a 125-bp region of DNA. We studied the assembly and function of kinetochore components by fusing individual kinetochore proteins to the lactose repressor (LacI) and testing their ability to improve segregation of a plasmid carrying tandem repeats of the lactose operator (LacO). Targeting Ask1, a member of the Dam1-DASH microtubule-binding complex, creates a synthetic kinetochore that performs many functions of a natural kinetochore: it can replace an endogenous kinetochore on a chromosome, bi-orient sister kinetochores at metaphase during the mitotic cycle, segregate sister chromatids, and repair errors in chromosome attachment. We show the synthetic kinetochore functions do not depend on the DNA-binding components of the natural kinetochore but do require other kinetochore proteins. We conclude that tethering a single kinetochore protein to DNA triggers assembly of the complex structure that directs mitotic chromosome segregation.
Figures
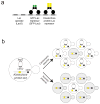
Figure 1. Assay for a synthetic kinetochore
A) Representation of binding of the Lac repressor (LacI) to the Lac operator (LacO). A dimer of LacI binds to palindromic LacO sequences. In our assays, we express a GFP-LacI fusion protein, a kinetochore protein-LacI fusion, or both (so that we can visualize the synthetic kinetochore). B) A diagram of the assay to determine if tethering individual proteins to DNA forms a synthetic kinetochore. Cells express a chromosomally integrated gene that fuses a kinetochore protein to LacI and the fusion protein is recruited to a plasmid that carries an array of tandem repeats of LacO and confers the ability to synthesize leucine, but lacks a centromere (acentric). The cells are grown in the absence of selection and the fraction containing the plasmid is measured. If the tethered protein confers kinetochore function, the plasmid will segregate to many of the daughter cells (like the control centromeric plasmid); if not, only a small fraction of the cells retain the plasmid (like the control acentric plasmid).
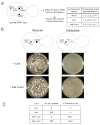
Figure 2. The synthetic kinetochore can replace a natural kinetochore
A) The presence of a synthetic kinetochore interferes with normal chromosome segregation. The cartoon shows the relevant features of the genotype of a diploid strain carrying one normal copy of chromosome III and one that carried both the natural and synthetic kinetochore. We measured the loss frequency of this chromosome in cells where the synthetic kinetochore is active (due to expression of the Ask1-LacI fusion) and those where it is not (cells without LacI and cells expressing unfused LacI). We first looked for loss of URA3 and then looked for those that had also lost LEU2 to determine that the entire chromosome was lost. The quoted range represents the 95% confidence interval. B) The synthetic kinetochore can substitute for a natural kinetochore. Cartoon of a conditional centromere. Galactose-induced transcription from the GAL1 promoter inactivates CEN3, rendering the natural kinetochore functional on glucose but inactive on galactose. Only cells expressing the Ask1-LacI fusion form colonies on galactose plates. Pictures were taken after 3 days of growth on glucose plates and 5 days of growth on galactose plates at 30°C. C) 8 or 256 repeats of the LacO array give similar results. Plating experiments were performed on strains with 8 or 256 repeats of the LacO array. Approximately 500 cells were plated to YPGlu or YPGal. The percent of cells that survived on YPGal compared to YPGlu was calculated and the average taken from 3 independent experiments where 4 plates were counted per experiment ± s.d.
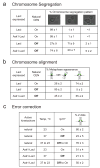
Figure 3. The synthetic kinetochore can align and segregate chromosomes and correct attachment errors
A) The synthetic kinetochore directs chromosome segregation. To monitor the segregation of sister chromatids, cells whose chromosome III carried both the LacO array and the conditional centromere (see Fig. 2B) were grown in either glucose (CEN3 ON) or galactose (CEN3 OFF) and arrested in anaphase. The synthetic kinetochore is active in cells expressing Ask1-LacI and inactive in those expressing LacI. All cells are expressing GFP-LacI and thus the sister chromatids are marked with GFP dots at the LacO array. Cells were classified into three categories: GFP dots separated with one in mother and one in the bud, two GFP dots in the mother, and two GFP dots in the daughter. Values in this figure represent the mean percent of cells ± s.d. from 3 independent experiments where at least 200 cells per experiment were counted. B) The synthetic kinetochore aligns chromosomes on the metaphase spindle. To determine if sister chromatids are able to biorient at metaphase, cells containing the LacO array and the conditional centromere were grown in either glucose or galactose, arrested in metaphase, and analyzed for transient separation of sister chromatids. If sister chromatids are transiently separated, two GFP dots are detected. If sister chromatids are bioriented but not separated or are mono-oriented, only one GFP dot is detected. The synthetic kinetochore is active in cells expressing Ask1-LacI and inactive in those expressing LacI. C). The synthetic kinetochore corrects errors in initial chromosome alignment. To verify that Ipl1 is needed for correct alignment of the synthetic kinetochore, cells carrying the temperature sensitive ipl1-321 mutant were arrested in G1 and then allowed to proceed to a metaphase arrest at either 23 °C (Ipl1 on), or 37 °C (Ipl1 off). To measure error correction, cells that had gone from G1 to the metaphase arrest at 37 °C were returned to 23 °C, restoring the activity of Ipl1. All assays with the synthetic kinetochore were conducted after inactivating the natural centromere and cells were scored as having one or two GFP-LacI dots.
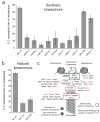
Figure 4. The synthetic kinetochore requires many components of natural kinetochores
A) Dissection of the genetic requirements for the synthetic kinetochore. Cells carrying temperature-sensitive mutations in a variety of kinetochore proteins were analyzed for their ability to biorient a chromosome carrying a synthetic kinetochore at 37 °C. The control (Syn. Kt.) contains the synthetic kinetochore but does not have any kinetochore temperature-sensitive mutations. Graphs in part A and B show the mean values ± s.d. of 3 experiments where at least 200 cells per genotype were counted per experiment. The values for the percent with 2 separated dots are the following: syn. kt. 55 ± 2, dam1-1 15 ± 5, ndc80-1 11 ± 6, nuf2-61 5 ± 3, spc24-1 5 ± 3, nsl1-54 14 ± 9, dsn1-7 7 ± 1, mtw1-1 11 ± 5, okp1-5 17 ± 11, ctf13-30 50 ± 3, ndc10-1 41 ± 5. B) Mutants that allow the synthetic kinetochore to biorient prevent biorientation of natural kinetochores. A natural kinetochore was analyzed for its ability to biorient with either an ndc10-1 or ctf13-30 temperature sensitive mutation. The wildtype control (wt kt) does not have any kinetochore temperature-sensitive mutations. The values for the percent with 2 separated dots are the following: wt. kt. 74 ± 1, ndc10-1 18 ± 1, ctf13-30 24 ± 4. C) A cartoon showing the location of proteins whose absence keeps the synthetic kinetochore from biorienting (shown in red). Proteins that are dispensable for the synthetic kinetochore are shown in blue, and proteins listed in black were not tested. Components of the DNA-binding complex are outlined in green. Components of the microtubule-binding complex are outlined in purple.
Comment in
- A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast.
Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann S. Kiermaier E, et al. Nat Cell Biol. 2009 Sep;11(9):1109-15. doi: 10.1038/ncb1924. Epub 2009 Aug 16. Nat Cell Biol. 2009. PMID: 19684577
Similar articles
- A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast.
Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann S. Kiermaier E, et al. Nat Cell Biol. 2009 Sep;11(9):1109-15. doi: 10.1038/ncb1924. Epub 2009 Aug 16. Nat Cell Biol. 2009. PMID: 19684577 - The phosphorylation of a kinetochore protein Dam1 by Aurora B/Ipl1 kinase promotes chromosome bipolar attachment in yeast.
Jin F, Bokros M, Wang Y. Jin F, et al. Sci Rep. 2017 Sep 19;7(1):11880. doi: 10.1038/s41598-017-12329-z. Sci Rep. 2017. PMID: 28928489 Free PMC article. - Molecular analysis of kinetochore-microtubule attachment in budding yeast.
He X, Rines DR, Espelin CW, Sorger PK. He X, et al. Cell. 2001 Jul 27;106(2):195-206. doi: 10.1016/s0092-8674(01)00438-x. Cell. 2001. PMID: 11511347 - The composition, functions, and regulation of the budding yeast kinetochore.
Biggins S. Biggins S. Genetics. 2013 Aug;194(4):817-46. doi: 10.1534/genetics.112.145276. Genetics. 2013. PMID: 23908374 Free PMC article. Review. - Structural view of the yeast Dam1 complex, a ring-shaped molecular coupler for the dynamic microtubule end.
Wu S, Grishchuk EL. Wu S, et al. Essays Biochem. 2020 Sep 4;64(2):359-370. doi: 10.1042/EBC20190079. Essays Biochem. 2020. PMID: 32579171 Free PMC article. Review.
Cited by
- The yeast 2-micron plasmid Rep2 protein has Rep1-independent partitioning function.
Mereshchuk A, Johnstone PS, Chew JSK, Dobson MJ. Mereshchuk A, et al. Nucleic Acids Res. 2022 Oct 14;50(18):10571-10585. doi: 10.1093/nar/gkac810. Nucleic Acids Res. 2022. PMID: 36156142 Free PMC article. - Centromere-like regions in the budding yeast genome.
Lefrançois P, Auerbach RK, Yellman CM, Roeder GS, Snyder M. Lefrançois P, et al. PLoS Genet. 2013;9(1):e1003209. doi: 10.1371/journal.pgen.1003209. Epub 2013 Jan 17. PLoS Genet. 2013. PMID: 23349633 Free PMC article. - Molecular underpinnings of centromere identity and maintenance.
Sekulic N, Black BE. Sekulic N, et al. Trends Biochem Sci. 2012 Jun;37(6):220-9. doi: 10.1016/j.tibs.2012.01.003. Epub 2012 Mar 10. Trends Biochem Sci. 2012. PMID: 22410197 Free PMC article. Review. - The selfish yeast plasmid exploits a SWI/SNF-type chromatin remodeling complex for hitchhiking on chromosomes and ensuring high-fidelity propagation.
Ma CH, Kumar D, Jayaram M, Ghosh SK, Iyer VR. Ma CH, et al. PLoS Genet. 2023 Oct 9;19(10):e1010986. doi: 10.1371/journal.pgen.1010986. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37812641 Free PMC article. - The Dam1 ring binds to the E-hook of tubulin and diffuses along the microtubule.
Ramey VH, Wang HW, Nakajima Y, Wong A, Liu J, Drubin D, Barnes G, Nogales E. Ramey VH, et al. Mol Biol Cell. 2011 Feb 15;22(4):457-66. doi: 10.1091/mbc.E10-10-0841. Epub 2010 Dec 17. Mol Biol Cell. 2011. PMID: 21169562 Free PMC article.
References
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. - PubMed
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. - PubMed
- Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. - PubMed
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous