Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling - PubMed (original) (raw)
Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling
Esra Cagavi Bozkulak et al. Mol Cell Biol. 2009 Nov.
Abstract
Notch signaling requires a series of proteolytic cleavage events to release the Notch intracellular domain (NICD) that functions directly in signal transduction. The Notch receptor is locked down in a protease-resistant state by a negative regulatory region (NRR) that protects an ADAM (a disintegrin and metalloprotease) cleavage site. Engagement with ligand-bearing cells induces global conformational movements in Notch that unfold the NRR structure to expose the ADAM cleavage site and initiate proteolytic activation. Although both ADAM10 and ADAM17 have been reported to cleave Notch to facilitate NICD release by gamma-secretase, the relevant ADAM has remained controversial. Our study provides new insight into this conflict, as we find that although Notch1 (N1) is a substrate for both ADAM10 and ADAM17, the particular ADAM required for receptor activation is context dependent. Specifically, ADAM10 was absolutely required for N1 signaling induced by ligands, while signaling independent of ligands required ADAM17. In contrast to the strict and differential use of ADAM10 and ADAM17 in normal and dysregulated signaling, respectively, both proteases participated in signaling intrinsic to N1 mutations associated with leukemia. We propose that in addition to exposing the ADAM cleavage site, activating N1 conformational changes facilitate selective cleavage by specific proteases.
Figures
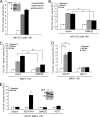
FIG. 1.
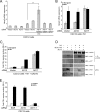
ADAM10 functions cell autonomously in Notch signaling. (A) ADAM10 is not required by Dll1 cells to activate signaling. NIH 3T3 cells coexpressing N1 and a CSL reporter were cocultured with stable Dll1HA-ADAM10+/+ (AD10+/+) or Dll1HA-ADAM10−/− (AD10−/−) MEFs and assayed for activation (activation over parental MEFs; n = 3). ADAM10 protein expression was determined for ADAM10+/+ and ADAM10−/− MEF ConA-enriched lysates by Western blotting (WB) (inset). (B and C) Ectopic expression of DNKUZ reduces ligand-induced Notch signaling. Reporter activity for NIH 3T3 cells (B) or wild-type MEFs (C) coexpressing DNKUZ with N1 induced by Dll1 or J1 cells and assayed for luciferase activity (activation over parental L cells; n = 4). (D) Ligand-induced Notch signaling is defective in ADAM10−/− MEFs. Dll1- or J1-induced reporter activity was measured in ADAM10−/− and ADAM10+/+ MEFs (activation over parental L cells; n = 6). (E) Ectopic ADAM10HA expression rescues ADAM10−/− MEF reporter activity. Reporter activity for ADAM10−/− MEFs cocultured with Dll1 cells following ectopic ADAM10HA (AD10-HA), DNKUZ, or ADAM17 (AD17) expression was measured as described for panel D (n = 4). ADAM17 protein expression was determined for ADAM10+/+ and ADAM10−/− MEF ConA-enriched lysates by Western blotting (inset). P, precursor form; M, mature; α, anti; **, P < 0.01; ***, P < 0.001.
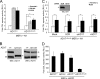
FIG. 2.
ADAM17 (AD17) is not required for ligand-induced Notch signaling (A) ADAM17ΔZn/ΔZn MEFs (AD17ΔZn/ΔZn) displayed a significant, but low level of reporter activity induced by ligand cells. Reporter activities for N1-expressing ADAM17ΔZn/ΔZn and ADAM17+/+ MEFs cocultured with Dll1 or J1 cells were assayed for luciferase activity (induction over parental L cells; n = 4). (B) ADAM17ΔZn/ΔZn MEFs express a targeted (T) ADAM17 protein and display ADAM10 (AD10) protein levels similar to those displayed by ADAM17+/+ MEFs. ADAM10 and ADAM17 protein expression was determined for ConA-enriched lysates isolated from ADAM17+/+ and ADAM17ΔZn/ΔZn MEFs by Western blotting (WB). (C) ADAM17ΔZn/ΔZn protein does not alter Notch signaling. ADAM17ΔZn/ΔZn MEFs treated with SCR or ADAM17 siRNA (siAD17) were cotransfected with ADAM17 and the CSL reporter constructs and then cocultured with Dll1 cells and assayed for luciferase activity (induction over parental L cells; n = 3). Specific siRNA depletion of ADAM17ΔZn/ΔZn protein was assayed by Western blotting (inset). (D) ADAM17ΔZn/ΔZn MEFs have intrinsically low Notch signaling activity. Reporter activity induced by constitutively active NICD in ADAM17ΔZn/ΔZn MEFs was assayed for luciferase activity (activation over vector transfected cells; n = 3). P, precursor form; M, mature; α, anti; *, P < 0.05.
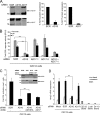
FIG. 3.
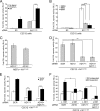
ADAM10, but not ADAM17, is required for ligand-induced Notch-dependent reporter activity and endogenous target gene (Herp2 gene) expression. (A) siRNA depletion of ADAM10 (AD10) and ADAM17 (AD17) proteins is specific and efficient. Western blot (WB) analysis and quantification of ADAM10 and ADAM17 proteins following siRNA depletion were performed (percentage relative to SCR-treated cells arbitrarily set to 100; representative of 10 experiments). (B) Depletion of ADAM10, but not ADAM17, decreased CSL reporter activity. C2C12 cells depleted for ADAM10 and/or ADAM17 protein by siRNA were transfected with the CSL reporter followed by coculture with Dll1 or J1 cells and assayed for luciferase activity (induction over parental L cells; n = 4). (C) Ectopic ADAM10HA (AD10HA or A10HA) expression rescued reporter activity in ADAM10 (AD10 or A10)-depleted C2C12 cells. Dll1-induced reporter activity was measured in C2C12 cells treated with SCR or ADAM10 siRNAs and then transfected with ADAM10 or ADAM17 (AD17 or A17) cDNAs (induction over parental L cells; n = 4). siRNA depletion and ectopic expression of ADAM10 and ADAM17 in ConA-enriched lysates were assayed by Western blotting (inset). (D) Depletion of ADAM10, but not ADAM17, decreased Dll1-induced endogenous Herp2 expression. C2C12 cells treated with indicated siRNAs were cocultured with Dll1 cells, and Herp2 expression was detected by qRT-PCR analysis (see Materials and Methods for details) (n = 4). P, precursor form; M, mature; α, anti; ***, P < 0.001.
FIG. 4.
ADAM10 expression is required for ligand-induced S2 cleavage of N1 in MEFs. (A) ADAM10−/− (AD10−/−) MEFs are defective in S2 cleavage. Dll1-induced N1Δmyc proteolytic fragments produced in stable ADAM10+/+ and ADAM10−/− MEFs treated with metalloprotease (BB94) or γ-secretase (DAPT) inhibitors (DMSO vehicle) were analyzed following immunoprecipitation (IP) and Western blotting (WB). The percent quantification of S1 fragment converted to S2 fragment from DAPT-treated Dll1 cocultures with ADAM10+/+ was arbitrarily set to 100% (lower panel; n = 4). (B) The residual S2 cleavage fragment detected with ADAM10−/− MEFs is not ADAM17 derived. The ligand-induced S2 cleavage fragment was analyzed by Western blotting in N1Δmyc ADAM10+/+ or ADAM10−/− MEFs following siRNA depletion of ADAM17 and coculture with Dll1 cells. The results shown are representative of one of two experiments. (C) Dll1-induced S2 cleavage is unaltered in ADAM17ΔZn/ΔZn MEFs. N1Δmyc-expressing ADAM17+/+ and ADAM17ΔZn/ΔZn MEFs were assayed and quantified as described for panel A (n = 4). *, P < 0.05.
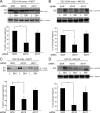
FIG. 5.
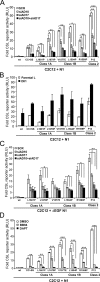
ADAM10, but not ADAM17, expression is required for ligand-induced S2 and S3 cleavage of N1Δmyc and endogenous N1 in C2C12 cells. (A and B) Depletion of ADAM10, but not ADAM17, decreased ligand-induced S2 and S3 cleavage of ectopic N1Δmyc. Dll1-induced S2 (A) and S3 (B) N1Δmyc cleavage fragments produced in ADAM10 (AD10) or ADAM17 (AD17) siRNA-depleted C2C12 cells in the presence of the γ-secretase (DAPT) (A) or proteasome (MG132) inhibitors (B) were detected by Western blotting (WB). The percent quantification of S1 fragment converted to S2 fragment from DAPT-treated Dll1 cocultures (A) and the percent quantification of S3 cleavage fragment in MG132-treated Dll1 cocultures (B) with SCR siRNA were arbitrarily set to 100% (lower panels; n = 4). (C and D) Depletion of ADAM10, but not ADAM17, decreased ligand-induced S2 and S3 cleavage of endogenous N1. Dll1-induced S2 (C) and S3 (D) endogenous N1 cleavage fragments produced in parental C2C12 cells were assayed and quantified as described for panels A and B, respectively. Lysates were immunoprecipitated (IP), and endogenous N1 proteolytic cleavage fragments were detected by Western blotting (n = 3). α, anti; *, P < 0.05; **, P < 0.01.
FIG. 6.
ADAM17 activates Notch signaling independent of ligand. (A) Overexpression of ADAM17 (AD17) with N1 induced ligand-independent Notch signaling that requires both ADAM and γ-secretase activity. C2C12 cells cotransfected with N1 and either ADAM10 (AD10) or ADAM17 (AD17) were incubated with metalloprotease (BB94) and γ-secretase (DAPT) inhibitors (DMSO vehicle), and Herp2 expression was detected by qRT-PCR analysis (see Materials and Methods for details) (n = 3). (B) Herp2 expression induced by ADAM17 is not sensitive to ligand. Herp2 mRNA was detected by qRT-PCR analysis in C2C12 cells, as described for panel A, and cocultured with Dll1 cells (n = 2). (C) ADAM10 is not required for reporter activity induced by ADAM17 independent of ligand. CSL reporter activity in ADAM10 siRNA depleted C2C12 cells overexpressing N1 with either ADAM10 or ADAM17 cDNAs was measured, following coculture with Dll1 cells (induction over parental L cells; n = 2). (D) ADAM17 interacts with ectopic but not endogenous N1. C2C12 cells expressing ADAM10 or ADAM17 either alone or together with N1 were either immunoblotted (WB) with anti-NICD (93-4), anti-AD10, or anti-AD17 antibodies or immunoprecipitated (IP) with anti-NICD (PCR 12), followed by WB with the indicated antibodies. The p120 furin-cleaved NICD fragment is shown for both NICD blots. (E) EDTA-induced reporter activity is ADAM17 dependent. C2C12 cells treated with the indicated siRNAs and expressing a CSL reporter were treated with either 1 mM EDTA or HBSS buffer for 10 min and assayed for luciferase activity 6 to 8 h later (n = 2). α, anti; **, P < 0.01.
FIG. 7.
ADAM17 is required for constitutive N1CC→SS signaling activity. (A) N1CC→SS reporter activity is enhanced by ligand. C2C12 cells coexpressing either N1 or N1CC→SS and a CSL reporter were assayed for activation in the presence and absence of Dll1 cells (activation over N1 cells in the absence of ligand; n = 3). (B) Constitutive N1CC→SS activity requires ADAM and γ-secretase cleavage. Reporter activity was measured for N1- or N1CC→SS-expressing C2C12 cells treated with metalloprotease (BB94) or γ-secretase (DAPT) inhibitors (activation over DMSO-treated N1 cells; n = 3). (C) ADAM10 (AD10) is not required for constitutive signaling by N1CC→SS. ADAM10+/+ or ADAM10−/− MEFs were cotransfected with either N1 or N1CC→SS and CSL reporter constructs and assayed for luciferase activity (activation over N1 cells; n = 3). (D) N1CC→SS constitutive activity requires ADAM17 (AD17). CSL reporter activity for N1CC→SS-expressing C2C12 cells depleted for ADAM10, ADAM17, or both by siRNAs and assayed for luciferase activity (activation over SCR siRNA-treated N1 cells; n = 3). (E) Losses in N1CC→SS reporter activity following ADAM17 or both ADAM10 and ADAM17 siRNA depletion can be rescued by ectopic expression of ADAM17 cDNA (activation over SCR-treated, vector-transfected N1 cells; n = 2). (F) N1CC→SS signaling induced by Dll1 is a composite of ligand-independent (open bars) and -dependent (filled bars) activation that requires both ADAM10 and ADAM17. Reporter activity of N1CC→SS-expressing C2C12 cells treated with ADAM10, ADAM17, or both siRNAs were cocultured with either L or Dll1 cells (activation over SCR-treated N1 cells cocultured with parental L cells; n = 3).
FIG. 8.
Signaling activity intrinsic to N1 T-ALL mutants have enhanced protease sensitivity. (A and C) T-ALL activating mutants require both ADAM10 and ADAM17 for constitutive signaling activity. C2C12 cells treated with ADAM10 (siAD10) and/or ADAM17 (siAD17) siRNAs were transfected with the indicated full-length (A) or EGF-truncated (ΔEGF) (C) class1 and 2 T-ALL N1 mutants and assayed for luciferase activity (activation over SCR-treated wild-type N1 cells; n = 2). (B) Signaling by T-ALL mutants can be enhanced by ligand. C2C12 cells cotransfected with the CSL reporter and the full-length N1 cDNAs with indicated mutations were cocultured with Dll1 cells (activation over wild-type N1 cells cocultured with L cells; n = 2). (D) Inhibition of γ-secretase but not metalloproteases efficiently suppressed signaling by class 1 T-ALL mutants. Reporter activity was detected for C2C12 cells expressing the indicated full-length N1 T-ALL mutants following treatment with metalloprotease (BB94) or γ-secretase (DAPT) inhibitors (DMSO vehicle) (activation over wild-type N1 cells treated with DMSO; n = 2).
Similar articles
- Analysis of the Conditions That Affect the Selective Processing of Endogenous Notch1 by ADAM10 and ADAM17.
Alabi RO, Lora J, Celen AB, Maretzky T, Blobel CP. Alabi RO, et al. Int J Mol Sci. 2021 Feb 12;22(4):1846. doi: 10.3390/ijms22041846. Int J Mol Sci. 2021. PMID: 33673337 Free PMC article. - A Disintegrin and Metalloproteinase10 (ADAM10) Regulates NOTCH Signaling during Early Retinal Development.
Toonen JA, Ronchetti A, Sidjanin DJ. Toonen JA, et al. PLoS One. 2016 May 25;11(5):e0156184. doi: 10.1371/journal.pone.0156184. eCollection 2016. PLoS One. 2016. PMID: 27224017 Free PMC article. - Metalloprotease ADAM10 is required for Notch1 site 2 cleavage.
van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. van Tetering G, et al. J Biol Chem. 2009 Nov 6;284(45):31018-27. doi: 10.1074/jbc.M109.006775. Epub 2009 Sep 2. J Biol Chem. 2009. PMID: 19726682 Free PMC article. - The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer.
Pruessmeyer J, Ludwig A. Pruessmeyer J, et al. Semin Cell Dev Biol. 2009 Apr;20(2):164-74. doi: 10.1016/j.semcdb.2008.09.005. Epub 2008 Sep 18. Semin Cell Dev Biol. 2009. PMID: 18951988 Review. - The "A Disintegrin And Metalloproteases" ADAM10 and ADAM17: novel drug targets with therapeutic potential?
Saftig P, Reiss K. Saftig P, et al. Eur J Cell Biol. 2011 Jun-Jul;90(6-7):527-35. doi: 10.1016/j.ejcb.2010.11.005. Epub 2010 Dec 30. Eur J Cell Biol. 2011. PMID: 21194787 Review.
Cited by
- Human NOTCH2 Is Resistant to Ligand-independent Activation by Metalloprotease Adam17.
Habets RA, Groot AJ, Yahyanejad S, Tiyanont K, Blacklow SC, Vooijs M. Habets RA, et al. J Biol Chem. 2015 Jun 5;290(23):14705-16. doi: 10.1074/jbc.M115.643676. Epub 2015 Apr 27. J Biol Chem. 2015. PMID: 25918160 Free PMC article. - ADAM-Mediated Signalling Pathways in Gastrointestinal Cancer Formation.
Schumacher N, Rose-John S, Schmidt-Arras D. Schumacher N, et al. Int J Mol Sci. 2020 Jul 20;21(14):5133. doi: 10.3390/ijms21145133. Int J Mol Sci. 2020. PMID: 32698506 Free PMC article. Review. - Analysis of the Conditions That Affect the Selective Processing of Endogenous Notch1 by ADAM10 and ADAM17.
Alabi RO, Lora J, Celen AB, Maretzky T, Blobel CP. Alabi RO, et al. Int J Mol Sci. 2021 Feb 12;22(4):1846. doi: 10.3390/ijms22041846. Int J Mol Sci. 2021. PMID: 33673337 Free PMC article. - Notch Signaling and Cross-Talk in Hypoxia: A Candidate Pathway for High-Altitude Adaptation.
O'Brien KA, Murray AJ, Simonson TS. O'Brien KA, et al. Life (Basel). 2022 Mar 16;12(3):437. doi: 10.3390/life12030437. Life (Basel). 2022. PMID: 35330188 Free PMC article. Review. - ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons.
Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. Kuhn PH, et al. EMBO J. 2010 Sep 1;29(17):3020-32. doi: 10.1038/emboj.2010.167. Epub 2010 Jul 30. EMBO J. 2010. PMID: 20676056 Free PMC article.
References
- Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. - PubMed
- Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. - PubMed
- Brou, C., F. Logeat, N. Gupta, C. Bessia, O. LeBail, J. R. Doedens, A. Cumano, P. Roux, R. A. Black, and A. Israel. 2000. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5:207-216. - PubMed
- Conlon, R. A., A. G. Reaume, and J. Rossant. 1995. Notch1 is required for the coordinate segmentation of somites. Development 121:1533-1545. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous