Hepatocyte growth factor family negatively regulates hepatic gluconeogenesis via induction of orphan nuclear receptor small heterodimer partner in primary hepatocytes - PubMed (original) (raw)
Hepatocyte growth factor family negatively regulates hepatic gluconeogenesis via induction of orphan nuclear receptor small heterodimer partner in primary hepatocytes
Dipanjan Chanda et al. J Biol Chem. 2009.
Abstract
Hepatic gluconeogenesis is tightly balanced by opposing stimulatory (glucagon) and inhibitory (insulin) signaling pathways. Hepatocyte growth factor (HGF) is a pleiotropic growth factor that mediates diverse biological processes. In this study, we investigated the effect of HGF and its family member, macrophage-stimulating factor (MSP), on hepatic gluconeogenesis in primary hepatocytes. HGF and MSP significantly repressed expression of the key hepatic gluconeogenic enzyme genes, phosphoenolpyruvate carboxykinase (PEPCK), and glucose-6-phosphatase (Glc-6-Pase) and reduced glucose production. HGF and MSP activated small heterodimer partner (SHP) gene promoter and induced SHP mRNA and protein levels, and the effect of HGF and MSP on SHP gene expression was demonstrated to be mediated via activation of the AMP-activated protein kinase (AMPK) signaling pathway. We demonstrated that upstream stimulatory factor-1 (USF-1) specifically mediated HGF effect on SHP gene expression, and inhibition of USF-1 by dominant negative USF-1 significantly abrogated HGF-mediated activation of the SHP promoter. Elucidation of the mechanism showed that USF-1 bound to E-box-1 in the SHP promoter, and HGF increased USF-1 DNA binding on the SHP promoter via AMPK and DNA-dependent protein kinase-mediated pathways. Adenoviral overexpression of USF-1 significantly repressed PEPCK and Glc-6-Pase gene expression and reduced glucose production. Knockdown of endogenous SHP expression significantly reversed this effect. Finally, knockdown of SHP or inhibition of AMPK signaling reversed the ability of HGF to suppress hepatocyte nuclear factor 4alpha-mediated up-regulation of PEPCK and Glc-6-Pase gene expression along with the HGF- and MSP-mediated suppression of gluconeogenesis. Overall, our results suggest a novel signaling pathway through HGF/AMPK/USF-1/SHP to inhibit hepatic gluconeogenesis.
Figures
FIGURE 1.
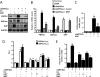
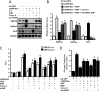
Inhibition of hepatic gluconeogenesis by HGF and MSP in primary hepatocytes. A–C, primary rat hepatocytes (A and B) and primary human hepatocytes (C) were pretreated with HGF (50 ng/ml, A and C), insulin (10 n
m
, 1 h), and MSP (100 ng/ml, B) for 3 h followed by treatment with cAMP (500 μ
m
) and Dex (100 n
m
) treatment for 3 h in the continuous presence or absence of HGF and MSP. Total RNA was isolated for Northern hybridization (A) and qPCR analysis (B and C). Data represent means ± S.D. of three individual experiments. *, p < 0.001 and **, p < 0.05 compared with untreated control and cAMP/Dex treated cells, respectively. D, AML12 cells were transfected with pepck and _Glc-6-Pase_-Luc (200 ng) for 24 h followed by treatment with cAMP (500 μ
m
) and Dex (100 n
m
) treatment for 3 h in the continuous presence or absence of HGF (50 and 100 ng/ml), MSP (100 and 200 ng/ml), or insulin as mentioned previously. Experiments were done in triplicate, and data are expressed in RLU and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments. *, p < 0.05 and **, p < 0.001 compared with untreated control and cAMP/Dex-treated cells, respectively. E, measurement of glucose production. Experiments were performed as described in A and B, using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 m
m
) and sodium pyruvate (1 m
m
). Data represent mean ± S.D. of four individual experiments. *, p < 0.001 and **, p < 0.001 compared with untreated control and cAMP/Dex treated cells, respectively. G6Pase, Glc-6-Pase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIGURE 2.
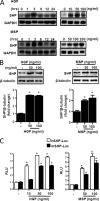
SHP gene expression is induced by HGF and MSP in primary hepatocytes. Primary hepatocytes were isolated from rats and cultured as described under “Experimental Procedures.” A, cells were treated with HGF or MSP at indicated concentrations or times, and total RNA was isolated for Northern hybridization. shp gene expression was analyzed and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression. The result is representative of three independently performed experiments. B, cells were treated with HGF or MSP at the indicated concentrations for 6 h and harvested for Western blot analysis using the indicated antibodies. The result shown is representative of three independently performed experiments. *, p < 0.001 compared with untreated control. C, HepG2 and AML12 cells were transfected with h_SHP_-Luc (200 ng) and m_Shp_-Luc (200 ng) respectively. 24 h after transfection, cells were serum-starved for a further 24 h, followed by HGF or MSP treatments at indicated concentrations for 12 h. Experiments were done in triplicate, and data are expressed in RLU, and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001 compared with untreated control.
FIGURE 3.
AMPK mediates HGF- and MSP-mediated induction of SHP gene expression. A, primary rat hepatocytes were pretreated with protein kinase inhibitors wortmannin (WM, 0.1 μ
m
), U0126 (U0, 10 μ
m
), SB203580 (SB, 25 μ
m
), SP600125 (SP, 25 μ
m
), compound C (C, 10 μ
m
) and H-89 (H89, 10 μ
m
) for 1 h followed by treatment with HGF for 3 h. Total RNA was isolated for semiquantitative RT-PCR analysis of shp mRNA expression and was normalized to β-actin expression. Data represent mean ± S.D. of three individual experiments. *, p < 0.05, and **, p < 0.001 compared with untreated control and HGF-treated cells. B, HepG2 and AML12 cells were transfected with h_SHP_-Luc (200 ng) and m_Shp_-Luc (200 ng), respectively. 24 h after transfection, cells were serum-starved for a further 24 h, followed by pretreatment of inhibitors for 1 h preceding HGF or MSP treatments at indicated concentrations for 12 h. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001, and **, p < 0.001 compared with untreated control and HGF- or MSP-treated cells, respectively. C, primary rat hepatocytes were treated with HGF or MSP at indicated concentration and times for 15 min (right) and harvested for Western blot analysis using indicated antibodies. Result shown is representative of three independently performed experiments. *, p < 0.001 compared with untreated control. D, PHHs were treated with HGF at indicated concentration and for indicated times (left) or treated with HGF for 3 h following pretreatment with compound C (Comp C) (10 μ
m
) (right). Cell lysates were extracted, and Western blot analysis was performed using the indicated antibodies (left). Total RNA was isolated for qPCR analysis SHP mRNA expression and was normalized to β-actin expression (right). Data represent mean ± S.D. of three individual experiments. *, p < 0.05, and **, p < 0.001 compared with untreated control and HGF-treated cells. E, primary rat hepatocytes (left and middle panels) were pretreated with compound C (10 μ
m
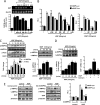
) for 1 h or infected with adenovirus dominant negative AMPKα (_Ad-dnAMPK_α) (50 m.o.i.) for 48 h followed by HGF treatment (50 ng/ml) and harvested for Western blot analysis using indicated antibodies. Result shown is representative of three independently performed experiments. HepG2 and AML12 cells were transfected (right panel) with h_SHP_-Luc (200 ng) and m_Shp_-Luc (200 ng) respectively, along with dnAMPKα expression vector (200 ng). 24 h after transfection, cells were serum-starved for a further 24 h, followed by HGF treatment (50 ng/ml) for 12 h. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001, and **, p < 0.001 compared with untreated control and HGF treated cells, respectively.
FIGURE 4.
USF-1 mediates HGF-mediated induction of SHP gene expression. A, HepG2 and AML12 cells were transfected with pcDNA3-FLAG-USF-1 (400 ng), pcDNA3-FLAG-USF-2 (400 ng), h_SHP_-Luc (200 ng), m_Shp_-Luc (200 ng), respectively (left), or HepG2 cells were transfected with h_SHP_-Luc (200 ng) (A middle and right, B and D), pcDNA3-FLAG-USF-1 (400 ng), and pcDNA3-FLAG-USF-2 (400 ng) (middle) or with pcDNA3-dnUSF-1 (400 ng) (right). 24 h post-serum starvation, cells were pretreated with compound C (comp C) (10 μ
m
) for 1 h preceding HGF (50 ng/ml) treatment. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001; **, p < 0.05; #, p < 0.001 compared with untreated control, USF-1 or HGF treated cells and HGF + USF-1 treated cells respectively. B and D, HepG2 cells were transfected with several deletion constructs (B) or E-box 1 mutant construct of hSHP-Luc (200 ng) (D) and USF-1 or treated with HGF as indicated. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001 compared with untreated control. C and E, ChIP assay. HepG2 cells were serum-starved for 24 h followed by HGF treatment for 12 h (C) or HepG2 cells were transfected with h_SHP_-Luc (−230/+1) (200 ng) wild type (wt) or E-box 1 mutant. Following transfection and serum starvation, cells were pretreated with compound C for 1 h preceding HGF treatment for 12 h. Soluble chromatin was prepared and immunoprecipitated with monoclonal antibody against USF-1 or IgG only as indicated. 10% of the soluble chromatin was used as input. qPCR was performed to determine and quantify the binding of USF-1 to endogenous (C) or transfected (E) h_SHP_ promoter. Data are representative of three individually performed experiments. *, p < 0.001, and **, p < 0.05 compared with untreated control and HGF treated cells respectively. ND, not detectable. F, HepG2 cells were cotransfected with pFR-Luc (200 ng), Gal4 constructs (400 ng each) containing DBD only, and USF-1, USF-1 T153A, and USF-1 S262A as indicated. Post-transfection and serum starvation, cells were pretreated with SB203580 or compound C following HGF treatment as indicated. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001, and **, p < 0.001 compared with untreated control and HGF-treated cells, respectively.
FIGURE 5.
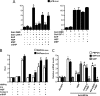
USF-1 represses gluconeogenesis via induction of SHP gene expression in primary hepatocytes. A and B, primary rat hepatocytes were infected with adenovirus (Ad) GFP (50 m.o.i.) or Ad-USF-1 (+ = 25 m.o.i., ++ = 50 m.o.i.) for 36 h. Cells were harvested for Western blot analysis using indicated antibodies (A), or total RNA was isolated for semi-quantitative RT-PCR analysis (B). Data represent mean ± S.D. of three individual experiments. *, p < 0.001 compared with Ad-GFP-treated cells. C, cells were infected with Ad-GFP, Ad-USF-1, or Ad-siSHP followed by Ad-USF-1 for 36–48 h preceding cAMP (500 μ
m
) and Dex (100 n
m
) treatment for 3 h. Total RNA was isolated for semiquantitative RT-PCR analysis. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.05, and #, p < 0.001 compared with Ad-GFP-infected cells, cAMP/Dex treatment, and Ad-USF-1 infected cells, respectively. D, measurement of glucose production. Experiments were performed as described in C, using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 m
m
) and sodium pyruvate (1 m
m
). Data represent mean ± S.D. of four individual experiments. *, p < 0.001; **, p < 0.001, and #, p < 0.001 compared with Ad-GFP infected cells, cAMP/Dex treatment, and Ad-USF-1 infected cells, respectively. G6Pase, Glc-6-Pase.
FIGURE 6.
HNF4α is a target of HGF-mediated repression of hepatic gluconeogenesis. A and B, HepG2 cells (A) and AML12 cells (B) were transfected with pFR-Luc, _pepck_-Luc, _Glc-6-Pase_-Luc (200 ng each), and expression vectors (400 ng each) as indicated. 24 h after transfection, cells were serum-starved and pretreated with compound C (comp C) (10 μ
m
) for 1 h (B) or treated with HGF (50 ng/ml) and MSP (100 ng/ml) for 12 h. Experiments were done in triplicate, and data are expressed RLU and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.001, and #, p < 0.001 compared with untreated control, Gal4-LRH-1- or HNF4α-transfected cells and HGF- or MSP-treated cells, respectively. C, primary hepatocytes were infected with Ad-GFP and Ad-HNF4α or infected with Ad-siSHP for 36–48 h or pretreated with compound C (10 μ
m
) for 1 h preceding HGF treatment for 3 h in the continuing presence of Ad-HNF4α. Total RNA was isolated for qPCR analysis. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.05, and #, p < 0.001 compared with Ad-GFP-infected cells, Ad-HNF4α alone- infected cells and HGF-treated cells, respectively. G6Pase, Glc-6-Pase.
FIGURE 7.
Knockdown of SHP expression abolished HGF and MSP effects on hepatic gluconeogenesis. A and B, primary rat hepatocytes were infected with Ad-GFP (50 m.o.i.) and Ad-siSHP (50 m.o.i.) for 48 h or pretreated with compound C (10 μ
m
) for 1 h followed by cAMP (500 μ
m
) and Dex (100 n
m
) treatment for 3 h in the continuous presence or absence of HGF (50 ng/ml), MSP (100 ng/ml), and insulin (10 n
m
) for 3 h. G6Pase, Glc-6-Pase. Total RNA was isolated for Northern hybridization (A) and qPCR analysis (B). Data represent mean ± S.D. of three individual experiments. *, p < 0.05, **, p < 0.05, and #, p < 0.001 compared with Ad-GFP-infected cells, cAMP/Dex treatment, and HGF- or MSP-treated cells, respectively. C, AML12 cells were transfected with _pepck_-Luc, _Glc-6-Pase_-Luc (200 ng each), and siSHP oligonucleotide (20 n
m
) for 24 h, followed by pretreatment with compound C (10 μ
m
) and treatment with cAMP (500 μ
m
) and Dex (100 n
m
) for 3 h in the continuous presence or absence of HGF (50 ng/ml), MSP (100 ng/ml), or insulin as mentioned previously. Experiments were done in triplicate, and data are expressed as RLU and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments. *, p < 0.001, **, p < 0.001, and #, p < 0.001 compared with untreated control, cAMP/Dex-treated cells, and HGF- or MSP-treated cells, respectively. D, measurement of glucose production. Experiments were performed as described in A and B, using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 m
m
) and sodium pyruvate (1 m
m
). Data represent mean ± S.D. of four individual experiments. *, p < 0.001, **, p < 0.001, and #, p < 0.001 compared with Ad-GFP infected cells, cAMP/Dex treatment, and HGF- or MSP-treated cells, respectively.
FIGURE 8.
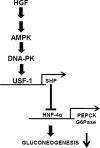
Schematic diagram of HGF-mediated inhibition of gluconeogenesis in primary hepatocytes. HGF signaling pathway activates AMPK signaling, leading to USF-1 activation via a DNA-PK-dependent pathway. USF-1 indirectly inhibits gluconeogenesis by binding to corepressor SHP gene promoter and induces SHP gene expression, which subsequently inhibits key gluconeogenic enzyme genes PEPCK and Glc-6-Pase (G6Pase) via inhibition of HNF4α and results in decreased gluconeogenesis.
Similar articles
- Orphan nuclear receptor small heterodimer partner negatively regulates growth hormone-mediated induction of hepatic gluconeogenesis through inhibition of signal transducer and activator of transcription 5 (STAT5) transactivation.
Kim YD, Li T, Ahn SW, Kim DK, Lee JM, Hwang SL, Kim YH, Lee CH, Lee IK, Chiang JY, Choi HS. Kim YD, et al. J Biol Chem. 2012 Oct 26;287(44):37098-108. doi: 10.1074/jbc.M112.339887. Epub 2012 Sep 12. J Biol Chem. 2012. PMID: 22977252 Free PMC article. - AMPK-dependent repression of hepatic gluconeogenesis via disruption of CREB.CRTC2 complex by orphan nuclear receptor small heterodimer partner.
Lee JM, Seo WY, Song KH, Chanda D, Kim YD, Kim DK, Lee MW, Ryu D, Kim YH, Noh JR, Lee CH, Chiang JY, Koo SH, Choi HS. Lee JM, et al. J Biol Chem. 2010 Oct 15;285(42):32182-91. doi: 10.1074/jbc.M110.134890. Epub 2010 Aug 5. J Biol Chem. 2010. PMID: 20688914 Free PMC article. - Sodium arsenite induces orphan nuclear receptor SHP gene expression via AMP-activated protein kinase to inhibit gluconeogenic enzyme gene expression.
Chanda D, Kim SJ, Lee IK, Shong M, Choi HS. Chanda D, et al. Am J Physiol Endocrinol Metab. 2008 Aug;295(2):E368-79. doi: 10.1152/ajpendo.00800.2007. Epub 2008 May 27. Am J Physiol Endocrinol Metab. 2008. PMID: 18505831 - Novel concepts in insulin regulation of hepatic gluconeogenesis.
Barthel A, Schmoll D. Barthel A, et al. Am J Physiol Endocrinol Metab. 2003 Oct;285(4):E685-92. doi: 10.1152/ajpendo.00253.2003. Am J Physiol Endocrinol Metab. 2003. PMID: 12959935 Review. - The Metformin Mechanism on Gluconeogenesis and AMPK Activation: The Metabolite Perspective.
Agius L, Ford BE, Chachra SS. Agius L, et al. Int J Mol Sci. 2020 May 3;21(9):3240. doi: 10.3390/ijms21093240. Int J Mol Sci. 2020. PMID: 32375255 Free PMC article. Review.
Cited by
- Quantitative Proteomic Analysis of Serum Reveals MST1 as a Potential Candidate Biomarker in Spontaneously Diabetic Cynomolgus Monkeys.
Tian C, Qiu M, Lv H, Yue F, Zhou F. Tian C, et al. ACS Omega. 2022 Dec 6;7(50):46702-46716. doi: 10.1021/acsomega.2c05663. eCollection 2022 Dec 20. ACS Omega. 2022. PMID: 36570245 Free PMC article. - Two polymorphisms of USF1 gene (-202G>A and -844C>T) may be associated with hepatocellular carcinoma susceptibility based on a case-control study in Chinese Han population.
Zhou X, Zhu HQ, Ma CQ, Li HG, Liu FF, Chang H, Lu J. Zhou X, et al. Med Oncol. 2014 Dec;31(12):301. doi: 10.1007/s12032-014-0301-4. Epub 2014 Nov 1. Med Oncol. 2014. PMID: 25367853 - Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective.
Wong RH, Sul HS. Wong RH, et al. Curr Opin Pharmacol. 2010 Dec;10(6):684-91. doi: 10.1016/j.coph.2010.08.004. Curr Opin Pharmacol. 2010. PMID: 20817607 Free PMC article. Review. - The role of AMPK-dependent pathways in cellular and molecular mechanisms of metformin: a new perspective for treatment and prevention of diseases.
Hasanvand A. Hasanvand A. Inflammopharmacology. 2022 Jun;30(3):775-788. doi: 10.1007/s10787-022-00980-6. Epub 2022 Apr 13. Inflammopharmacology. 2022. PMID: 35419709 Free PMC article. Review. - Regulation of Energy Metabolism by Receptor Tyrosine Kinase Ligands.
Zhao M, Jung Y, Jiang Z, Svensson KJ. Zhao M, et al. Front Physiol. 2020 Apr 21;11:354. doi: 10.3389/fphys.2020.00354. eCollection 2020. Front Physiol. 2020. PMID: 32372975 Free PMC article. Review.
References
- Desvergne B., Michalik L., Wahli W. (2006) Physiol. Rev. 86, 465–514 - PubMed
- Accili D. (2004) Diabetes 53, 1633–1642 - PubMed
- Song K. H., Ellis E., Strom S., Chiang J. Y. (2007) Hepatology 46, 1993–2002 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK44442/DK/NIDDK NIH HHS/United States
- R56 DK044442/DK/NIDDK NIH HHS/United States
- R01 DK058379/DK/NIDDK NIH HHS/United States
- R37 DK058379/DK/NIDDK NIH HHS/United States
- DK58379/DK/NIDDK NIH HHS/United States
- R01 DK044442/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials