Metalloprotease ADAM10 is required for Notch1 site 2 cleavage - PubMed (original) (raw)
Metalloprotease ADAM10 is required for Notch1 site 2 cleavage
Geert van Tetering et al. J Biol Chem. 2009.
Erratum in
- J Biol Chem. 2010 Apr 9;285(15):11754
Abstract
Notch signaling is controlled by ligand binding, which unfolds a negative control region to induce proteolytic cleavage of the receptor. First, a membrane-proximal cleavage is executed by a metalloprotease, removing the extracellular domain. This allows gamma-secretase to execute a second cleavage within the Notch transmembrane domain, which releases the intracellular domain to enter the nucleus. Here we show that the ADAM10 metalloprotease Kuzbanian, but not ADAM17/tumor necrosis factor alpha-converting enzyme, plays an essential role in executing ligand-induced extracellular cleavage at site 2 (S2) in cells and localizes this step to the plasma membrane. Importantly, genetic or pharmacological inhibition of metalloproteases still allowed extracellular cleavage of Notch, indicating the presence of unknown proteases with the ability to cleave at S2. Gain of function mutations identified in human cancers and in model organisms that map to the negative control region alleviate the requirement for ligand binding for extracellular cleavage to occur. Because cancer-causing Notch1 mutations also depend on (rate-limiting) S2 proteolysis, the identity of these alternative proteases has important implications for understanding Notch activation in normal and cancer cells.
Figures
FIGURE 1.
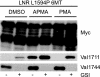
A, diagram depicting S1, S2, and S3 cleavage steps leading to NICD production and activity; see text for details. Boxed area indicates immunization peptide sequence. B, immunoblot showing expression of wild type (WT) and T-ALL mutant (L1594P) LNR 6Myc proteins transfected in HEK293 cells. Upper panel, anti-Myc immunoblot showing equal expression levels of transfected constructs. S1, S2, and S3 cleavage products are indicated. Lower panel, Val1744 immunoblot for S3-cleaved Notch1 showing NICD formation in L1594P but not in the inactive wild type. NICD production and Val1744 staining is blocked by inhibition of γ-secretase by GSIs DAPT and dibenzazepine. Middle panels, accumulation of S2-cleaved Notch1 detected by Val1711 only seen upon GSI treatment and concomitant loss of Val1744/NICD in panel below. Val1711 only observed in active L1594P mutant and not in wild type. Note accumulation of S2-cleaved Notch1 fragments is also observed in Myc immunoblot for L1594P upon GSI treatment. Incubation with the immunization peptide (N1) prevents detection of S2-cleaved Notch1 by Val1711, and control peptide against hNotch2 (N2) does not block immunoreactivity. C, Notch-CSL transcription reporter assay in U2OS cells showing wild type LNR 6Myc compared with LNR L1594P 6Myc, which is 5-fold more active. Notch1 activity is attenuated using GSI. TMIC, transmembrane and intracellular domain; DBZ, dibenzazepine.
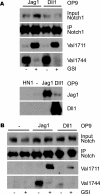
FIGURE 2.
Ligand-induced Notch1 signaling leads to Val1711 S2 cleavage. A, co-culture of wild type-, Jagged1- (Jag1), and Delta1 (Dll1)-expressing murine OP9 cells with mouse Notch1-expressing HeLaN1 (HN1). Immunoprecipitation (IP) for Notch1 shows Notch1 receptor expression. Immunoblot for Val1744 and Val1711 shows that both Jag1 and Dll1 induce NICD, which is blocked by GSIs. Both Dll1 and Jag1 induce S2 cleavage at Val1711 seen upon GSI treatment. Lower panel, expression of Jagged1 and Delta1 in OP9-transduced cells. No ligand expression is observed in HN1 cells. B, activation of endogenous Notch1 signaling by ligand stimulation in OP9 cells proceeds through Val1711 proteolysis. Immunoprecipitation shows OP9 ligand-expressing cells also express endogenous Notch1 receptor. OP9-Jag1 and OP9-Dll1 cultures undergo ligand-dependent Notch1 signaling to produce S2-cleaved Notch1 at Val1711. OP9 cells not expressing ligand do not activate Notch1 cleavage at Val1711 or Val1744.
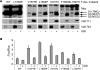
FIGURE 3.
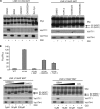
T-ALL ligand-independent Notch1 LNR molecules are cleaved at Val1711. A, upper panel, T-ALL LNR 6Myc HD mutants transfected in HEK293 cells. S1, S2, and S3 cleavage products are indicated. Lower panel, Val1744 immunoblot shows all T-ALL mutants but not wild type (WT) LNR produce NICD, which is inhibited by GSI. Middle panel shows Val1711 cleavage in T-ALL mutants but not wild type upon GSI treatment. B, transcriptional reporter activity of LNR T-ALL mutants on CSL-fLuc reporter in HeLa cells. Shown is fold Firefly luciferase (Fluc) activity corrected for Renilla luciferase (Rluc) transfection of LNR mutants over background. Note that the less efficient S2 and S3 cleavage of F1593S and L1597H in A is also reflected in the reduced activity in the CSL-fluc reporter assay. Luciferase assays are representative of at least two independent experiments in triplicate; all mutants are significantly (p < 0.05) more active compared with wild type. TMIC, transmembrane and intracellular domain.
FIGURE 4.
Val1711 cleavage site mutation leads to ligand-independent Notch1 cleavage and activity. A, Myc immunoblot of HEK293 cell lysates transfected with wild type (WT), S2 cleavage mutant (AV → VH), and L1594P full-length Notch1 6Myc constructs. Receptor dissociation and cleavage stimulated by EDTA induce Val1744 and Val1711 cleavage in wild type and L1594P but not in AV → VH S2 cleavage mutant. L1594P and AV → VH mutants are already Val1744-cleaved in the absence of EDTA. Val1711 cleavage in AV → VH cannot be monitored because of mutation of epitope. B, Notch1-GV16 cleavage reporter assay in HeLa cells showing EDTA-induced cleavage of WT, AV → VH, and L1594P mutants. AV → VH and L1594P are already highly active in the absence of EDTA compared with wild type. C, Notch1-GV16 cleavage assay in HeLa cells co-cultured with OP9 wild type or Jagged1 cells showing that the basal activity of AV → VH mutant and L1594P T-ALL mutant in absence of ligand is severalfold higher than wild type. Whereas the activity of the AV → VH mutant can be stimulated with Jagged1, L1594P T-ALL is not further stimulated. Shown is fold Firefly luciferase (Fluc) activity corrected for Renilla luciferase (Rluc) transfection of Notch1 6Myc constructs over background. Figure is representative of at least two independent experiments in triplicate; p values are shown and calculated using a Student's t test.
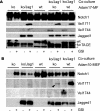
FIGURE 5.
ADAM10 is essential for endogenous Notch1 Val1711 cleavage. _A, Adam17_-deficient cells are transduced with Jagged1 ligand and used in co-culture experiments to stimulate wild type (WT) and _Adam17_-deficient cells that express endogenous Notch1 receptor. Immunoblotting for Notch1, Val1711, and Val1744 shows no defect is observed in Notch1 cleavage in the absence of ADAM17. ko, knock-out. B, in contrast in cells lacking ADAM10-expressing endogenous Notch1 receptor, S2 and S3 cleavages are severely impaired compared with wild type cells. Jagged1 and TACE/ADAM17 expression is shown. Asterisk indicates nonspecific reaction of TACE antibody.
FIGURE 6.
Constitutive and regulated cleavage of Notch1 at S2. HEK293 cells transfected with LNR L1594P 6Myc. Upper panel, Myc immunoblot. Lower panel shows that the phorbol ester PMA and mercuric compound APMA stimulate constitutive Val1711 and Val1744 cleavage compared with vehicle-treated cells. Note Val1711 cleavage is observed even in the absence of GSI with APMA, indicating Val1711 cleavage is not indirectly caused by GSI.
FIGURE 7.
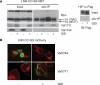
Notch1 S2 cleavage occurs at the cell surface. A, upper panel, Myc immunoblot of surface-biotinylated and streptavidin-precipitated U2OS cells transfected with the active LNR CC → SS 6Myc. Left upper panel shows input, and right panel streptavidin pulldown (sAv-IP) of biotinylated cells. Lower panels show corresponding Val1711 and Val1744 immunoblots. Streptavidin pulldown demonstrates S2-cleaved fragments enriched at the cell surface compared with Val1744-cleaved Notch present in input but not detected by surface biotinylation. Streptavidin pulldown on U2OS cells transfected with FLAG-tagged HIF1α protein shows biotin did not label cytoplasmic proteins. B, U2OS cells transfected with LNR CC → SS mCherry are fixed and permeabilized and used for immunofluorescent staining for Val1711- and Val1744-cleaved Notch1 (green). Val1744 staining is only present in absence of GSI, whereas Val1711-cleaved molecules are only present in GSI-treated cells predominantly located at the cell membrane. Red fluorescence shows total Notch1 expression. Val1711-positive vesicular structures are observed near the cell surface. TMIC, transmembrane and intracellular domain.
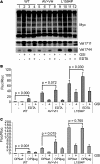
FIGURE 8.
Metalloprotease inhibitors block Val1711 cleavage but not S2 cleavage. A, broad spectrum metalloprotease inhibitors and ADAM-specific inhibitors show complete inhibition of Val1711 cleavage but not of Val1744-NICD production in LNR L1594P 6Myc-transfected HEK293 cells. The ADAM10-specific inhibitor GI254023 shows minimal inhibition of Val1711 cleavage (arrowhead). Serine-cysteine protease inhibitor mixture (Ser-Cys) does not affect S2 or S3 cleavage. B, Notch1-GV16 reporter assay in transfected NIH-3T3 cells showing only partial inhibition of Notch1 S3 cleavage measured by GV16 release with MPi GM6001 and TAPI-2 on constitutively active CC → SS constructs. Shown is fold Firefly luciferase (Fluc) activity corrected for Renilla luciferase (Rluc) transfection of Notch1 wild type and CC → SS-GV16 constructs over background. C, increased concentrations of MPi lead to a reduction of Val1744-NICD production. Note in Myc immunoblots S2-like products still accumulate in GSI- and MPi-treated cells, indicating that S2 cleavage occurs elsewhere when Val1711 cleavage is blocked. TMIC, transmembrane and intracellular domain; DBZ, dibenzazepine.
Similar articles
- Analysis of the Conditions That Affect the Selective Processing of Endogenous Notch1 by ADAM10 and ADAM17.
Alabi RO, Lora J, Celen AB, Maretzky T, Blobel CP. Alabi RO, et al. Int J Mol Sci. 2021 Feb 12;22(4):1846. doi: 10.3390/ijms22041846. Int J Mol Sci. 2021. PMID: 33673337 Free PMC article. - Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling.
Bozkulak EC, Weinmaster G. Bozkulak EC, et al. Mol Cell Biol. 2009 Nov;29(21):5679-95. doi: 10.1128/MCB.00406-09. Epub 2009 Aug 24. Mol Cell Biol. 2009. PMID: 19704010 Free PMC article. - Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and γ-secretase.
Hogl S, Kuhn PH, Colombo A, Lichtenthaler SF. Hogl S, et al. PLoS One. 2011;6(6):e21337. doi: 10.1371/journal.pone.0021337. Epub 2011 Jun 17. PLoS One. 2011. PMID: 21695060 Free PMC article. - Alpha-secretase cleavage of the amyloid precursor protein: proteolysis regulated by signaling pathways and protein trafficking.
Lichtenthaler SF. Lichtenthaler SF. Curr Alzheimer Res. 2012 Feb;9(2):165-77. doi: 10.2174/156720512799361655. Curr Alzheimer Res. 2012. PMID: 21605033 Review. - Implication of APP secretases in notch signaling.
Hartmann D, Tournoy J, Saftig P, Annaert W, De Strooper B. Hartmann D, et al. J Mol Neurosci. 2001 Oct;17(2):171-81. doi: 10.1385/JMN:17:2:171. J Mol Neurosci. 2001. PMID: 11816790 Review.
Cited by
- Targeting the Notch Signaling Pathway in Chronic Inflammatory Diseases.
Christopoulos PF, Gjølberg TT, Krüger S, Haraldsen G, Andersen JT, Sundlisæter E. Christopoulos PF, et al. Front Immunol. 2021 Apr 12;12:668207. doi: 10.3389/fimmu.2021.668207. eCollection 2021. Front Immunol. 2021. PMID: 33912195 Free PMC article. Review. - Conditional deletion of Notch1 and Notch2 genes in excitatory neurons of postnatal forebrain does not cause neurodegeneration or reduction of Notch mRNAs and proteins.
Zheng J, Watanabe H, Wines-Samuelson M, Zhao H, Gridley T, Kopan R, Shen J. Zheng J, et al. J Biol Chem. 2012 Jun 8;287(24):20356-68. doi: 10.1074/jbc.M112.349738. Epub 2012 Apr 13. J Biol Chem. 2012. PMID: 22505716 Free PMC article. - A Disintegrin and Metalloprotease (ADAM): Historical Overview of Their Functions.
Giebeler N, Zigrino P. Giebeler N, et al. Toxins (Basel). 2016 Apr 23;8(4):122. doi: 10.3390/toxins8040122. Toxins (Basel). 2016. PMID: 27120619 Free PMC article. Review. - ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo.
Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, Conrad DH. Gibb DR, et al. J Exp Med. 2010 Mar 15;207(3):623-35. doi: 10.1084/jem.20091990. Epub 2010 Feb 15. J Exp Med. 2010. PMID: 20156974 Free PMC article. - Rare mutations of ADAM17 from TOFs induce hypertrophy in human embryonic stem cell-derived cardiomyocytes via HB-EGF signaling.
Xie Y, Ma A, Wang B, Peng R, Jing Y, Wang D, Finnell RH, Qiao B, Wang Y, Wang H, Zheng Y. Xie Y, et al. Clin Sci (Lond). 2019 Jan 22;133(2):225-238. doi: 10.1042/CS20180842. Print 2019 Jan 31. Clin Sci (Lond). 2019. PMID: 30610007 Free PMC article.
References
- Wilson A., Radtke F. (2006) FEBS Lett. 580, 2860–2868 - PubMed
- Gordon W. R., Vardar-Ulu D., Histen G., Sanchez-Irizarry C., Aster J. C., Blacklow S. C. (2007) Nat. Struct. Mol. Biol. 14, 295–300 - PubMed
- Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) Mol. Cell 5, 197–206 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG013956/AG/NIA NIH HHS/United States
- GM55479/GM/NIGMS NIH HHS/United States
- R37 AG013956/AG/NIA NIH HHS/United States
- R01 GM055479/GM/NIGMS NIH HHS/United States
- 208259/ERC_/European Research Council/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous