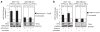miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells - PubMed (original) (raw)
doi: 10.1038/cdd.2009.117. Epub 2009 Sep 4.
M S Nicoloso, L Lupini, Y Lu, J Fogarty, S Rossi, B Zagatti, M Fabbri, A Veronese, X Liu, R Davuluri, C M Croce, G Mills, M Negrini, G A Calin
Affiliations
- PMID: 19730444
- PMCID: PMC3648637
- DOI: 10.1038/cdd.2009.117
miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells
R Spizzo et al. Cell Death Differ. 2010 Feb.
Abstract
Understanding the consequences of miR-145 reintroduction in human breast cancer (BC) could reveal its tumor-suppressive functions and may disclose new aspects of BC biology. Therefore, we characterized the effects of miR-145 re-expression in BC cell lines by using proliferation and apoptosis assays. As a result, we found that miR-145 exhibited a pro-apoptotic effect, which is dependent on TP53 activation, and that TP53 activation can, in turn, stimulate miR-145 expression, thus establishing a death-promoting loop between miR-145 and TP53. We also found that miR-145 can downregulate estrogen receptor-alpha (ER-alpha) protein expression through direct interaction with two complementary sites within its coding sequence. In conclusion, we described a tumor suppression function of miR-145 in BC cell lines, and we linked miR-145 to TP53 and ER-alpha. Moreover, our findings support a view that miR-145 re-expression therapy could be mainly envisioned in the specific group of patients with ER-alpha-positive and/or TP53 wild-type tumors.
Figures
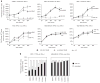
Figure 1
miR-145 inhibits breast cancer cell growth. (a) Assessment of cell number by tetrazolium staining (MTT assay) in six BC cell lines. MiR-145 effects on cell counting were measured up to 96 h after miR-145 transfection (time 0). Each time point was expressed as a relative value (fold change, Y axis) to time 0. Values represent the average and bars represent S.D. of three (MCF7 and MDA-MB-231) or two (all other cell lines) independent experiments carried out each time in quadruplicates. For each cell line, TP53 and ER-α protein status are shown according to Neve et al.42 For TP53, − indicates no expression, +/− low expression, + medium expression, ++ high expression, mut mutated and wt wild type. For ER-α, protein levels are indicated as + (positive) or − (negative). (b) Assessment of cell number by tetrazolium staining (MTT assay) of MCF-7 and MDA-MB-231 cell lines transfected with scalar amounts of miR-145 and scrambled (from 100 nM to 3.125 nM). The cells were counted 48 h after miR-145 and scrambled transfection. For each concentration, miR-145 effect was expressed as a relative value (fold change, Y axis) to scrambled control. The symbol asterisk represents a significant difference (P<0.05) compared with scrambled by t-test. Null cells were treated only with lipofectamine.
Figure 2
miR-145 induces apoptosis in BC cell lines. (a) Annexin V staining of BC cell lines at different time points after miR-145 transfection (time 0). For each time point, we measured the percentage of annexin V-positive and propidium iodide-negative cells (annexin V+/PI−) (Y axis). Values represent averages and bars represent S.D. of two independent experiments. (b) Detection of caspase 3/7 activity by luminescent assay (luminescence, Y axis) 24 and 48 h after miR-145 transfection. Cisplatin treatment (40 μM) was used as a control of apoptosis in MDA-MB-231. Values represent average and bars represent S.D. of three replicates. (c) Western blotting of PARP protein in four BC cell lines 24 and 48 h after miR-145 transfection. Two bands are shown, full-length PARP (116 kDa) and cleaved PARP (89 kDa). The cleaved form is the marker of apoptosis. An increased ratio between cleaved and total PARP indicates an induction of apoptosis. (d) miR-145 effects on caspase 3 and 7 activity were measured by luminescent assay (luminescence, Y axis) in three BC cell lines that were transfected with scalar concentrations of miR-145 (100 nM, 50 nM and 5 nM). (e) Western blotting of full-length and cleaved PARP in three BC cell lines after transfection with different concentrations of miR-145 (100 nM, 50 nM and 5 nM). The asterisk represents a statistically significant difference (P<0.05) compared with scrambled by t-test. Null cells were treated only with lipofectamine.
Figure 3
miR-145 enhances TP53 transcriptional activity and functions in part through TP53 pathway activation. (a) mRNA levels of PUMA and p21 measured by qRT-PCR 48 h after miR-145 or scrambled transfection in two cell lines, MCF7 (TP53 wt) and MDA-MB-231 (TP53 mutated). Fold changes of P21 and PUMA have been calculated using 2−ΔCt method. GAPDH mRNA levels were used as an internal normalization control. Values represent averages and bars represent S.D. of three independent experiments. Samples treated with miR-145 and scrambled have been normalized to null samples. (b) Western blotting for the indicated proteins in MCF-10 samples 24 h after transfection with miR-145 (100 nM) or scrambled (100 nM). GAPDH was used as loading controls. (c) P21 and PUMA mRNA levels measured by qRT-PCR after transfection of miR-145 and siRNA anti-TP53 or miR-145 alone in MCF-10A. Fold changes of P21 and PUMA have been calculated using 2−ΔCt method. GAPDH mRNA levels were used as an internal normalization control. The cells were first transfected with siRNA anti-TP53, and 36 h later the cells were transfected with miR-145, scrambled or with lipofectamine only (null). (d) Assessment of cell number by MTT assay in MCF-7 48 h after miR-145 transfection in the presence or absence (by siRNA) of TP53 or of PUMA. Y axis represent fold change relative to null cells. Values represent averages and bars represent S.D. of three experiments. At the bottom of the graph, western blottings of PUMA and β-actin are shown. The asterisk represents a statistically significant difference (P<0.05) compared with scrambled by t-test allowing unequal variances between samples. Null cells were treated only with lipofectamine.
Figure 4
TP53 induces miR-145 expression levels. (a and b) miR-145 and miR-34c levels were measured by qRT-PCR 24 h after treatment with Adriamycin and Nutlin-3, respectively. Fold changes of miR-145 and miR-34c have been calculated using 2−ΔCt method. U6 mRNA levels were used as an internal normalization control. Values represent average and bars represent S.D. of three independent experiments. At the bottom of the graph, western blotting of TP53 protein levels is shown to prove induction of TP53 protein levels after Adriamycin and Nutlin-3 treatment. GAPDH and β-actin protein levels were used as loading controls. The asterisk represents a significant difference (P<0.05) compared with untreated samples by t-test.
Figure 5
miR-145 represses ER-α protein in MCF-7 by direct binding within the ESR1-coding sequence. (a) Western blotting analysis of ER-α, cyclin D1, and phosphorylated ER-α in serine 118 (pS118) protein levels in MCF7 48 h after miR-145 transfection. GAPDH was used as loading control. (b) ER-α and cyclin D1 protein levels were measured by western blotting 48 h after miR-145 (100 nM) or scrambled transfection (100 nM). Values represent average and bars represent S.D. of three independent experiments, one of which is shown in panel a. GAPDH was used as internal normalization control. (c) ER-α protein levels were measured 48 h after transfection of scalar doses of miR-145 (100 nM, 50 nM and 5 nM). Vinculin was used as loading control. (d) Top part of the panel represents the isoform1 of ESR1 mRNA. The miR-145-predicted target sites are shown as triangles (white triangles if they are located in the 3′ UTR, black if they are located in the CDS), and they are labeled from 1 to 5 starting from the 5′ of the mRNA. The middle cartoon represents the luciferase construct that has been used for in vitro assay. The bottom panel represents the results of in vitro experiments with luciferase constructs that contain miR-145-predicted targets. Mutated constructs have been generated by deletion of seven nucleotides in the 3′ of the target sites. Values represent averages and bars represent S.D. of three independent experiments. Luciferase activity was normalized to that of scrambled control. The asterisk represents a statistically significant difference (P<0.05) compared with scrambled by t-test. Null cells were treated only with lipofectamine.
Figure 6
miR-145 does not decrease ESR1 mRNA or ER-α protein stability. (a) ESR1 mRNA and (b) ER-α pS118/ER-α protein levels were measured 48 h after miR-145 transfection. Values represent average and bars represent S.D. of three independent experiments. Fold changes of ESR1 mRNA levels were calculated using 2−ΔCt method. GAPDH mRNA levels were used as an internal normalization control. Fold changes of ER-α pS118/ER-α ratios were calculated by western blotting (see Figure 5a). The asterisk represents a significant difference (P<0.05) compared with scrambled by t-test. Null cells were treated only with lipofectamine.
Similar articles
- The ERα-miR-575-p27 feedback loop regulates tamoxifen sensitivity in ER-positive Breast Cancer.
Liu SS, Li Y, Zhang H, Zhang D, Zhang XB, Wang X, Yu Y. Liu SS, et al. Theranostics. 2020 Aug 29;10(23):10729-10742. doi: 10.7150/thno.46297. eCollection 2020. Theranostics. 2020. PMID: 32929377 Free PMC article. - Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status.
Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, Cheung DG, Bolon B, Anderlucci L, Alder H, Nuovo G, Li M, Iorio MV, Galasso M, Santhanam R, Marcucci G, Perrotti D, Powell KA, Bratasz A, Garofalo M, Nephew KP, Croce CM. Di Leva G, et al. PLoS Genet. 2013;9(3):e1003311. doi: 10.1371/journal.pgen.1003311. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505378 Free PMC article. - Estrogen levels act as a rheostat on p53 levels and modulate p53-dependent responses in breast cancer cell lines.
Fernández-Cuesta L, Anaganti S, Hainaut P, Olivier M. Fernández-Cuesta L, et al. Breast Cancer Res Treat. 2011 Jan;125(1):35-42. doi: 10.1007/s10549-010-0819-x. Epub 2010 Mar 11. Breast Cancer Res Treat. 2011. PMID: 20221692 - Estrogen regulates miRNA expression: implication of estrogen receptor and miR-124/AKT2 in tumor growth and angiogenesis.
Jiang CF, Li DM, Shi ZM, Wang L, Liu MM, Ge X, Liu X, Qian YC, Wen YY, Zhen LL, Lin J, Liu LZ, Jiang BH. Jiang CF, et al. Oncotarget. 2016 Jun 14;7(24):36940-36955. doi: 10.18632/oncotarget.9230. Oncotarget. 2016. PMID: 27175587 Free PMC article. - p53 in breast cancer subtypes and new insights into response to chemotherapy.
Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R, Turpin E, Plassa LF, de Roquancourt A, Bourstyn E, de Cremoux P, Janin A, Giacchetti S, Espié M, de Thé H. Bertheau P, et al. Breast. 2013 Aug;22 Suppl 2:S27-9. doi: 10.1016/j.breast.2013.07.005. Breast. 2013. PMID: 24074787 Review.
Cited by
- MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients.
Doberstein K, Steinmeyer N, Hartmetz AK, Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R, Pfeilschifter J, Gutwein P. Doberstein K, et al. Neoplasia. 2013 Feb;15(2):218-30. doi: 10.1593/neo.121222. Neoplasia. 2013. PMID: 23441135 Free PMC article. - MiRNA-145 increases therapeutic sensibility to gemcitabine treatment of pancreatic adenocarcinoma cells.
Lin Y, Ge X, Wen Y, Shi ZM, Chen QD, Wang M, Liu LZ, Jiang BH, Lu Y. Lin Y, et al. Oncotarget. 2016 Oct 25;7(43):70857-70868. doi: 10.18632/oncotarget.12268. Oncotarget. 2016. PMID: 27765914 Free PMC article. - MicroRNA-145 and MicroRNA-133a Inhibited Proliferation, Migration, and Invasion, While Promoted Apoptosis in Hepatocellular Carcinoma Cells Via Targeting FSCN1.
Wang G, Zhu S, Gu Y, Chen Q, Liu X, Fu H. Wang G, et al. Dig Dis Sci. 2015 Oct;60(10):3044-52. doi: 10.1007/s10620-015-3706-9. Epub 2015 Jul 15. Dig Dis Sci. 2015. PMID: 26173501 - MiR-145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c-Myc and Cdk6.
Shao Y, Qu Y, Dang S, Yao B, Ji M. Shao Y, et al. Cancer Cell Int. 2013 May 28;13(1):51. doi: 10.1186/1475-2867-13-51. Cancer Cell Int. 2013. PMID: 23710609 Free PMC article. - Inflammation related miRNAs as an important player between obesity and cancers.
Gholami M, Larijani B, Zahedi Z, Mahmoudian F, Bahrami S, Omran SP, Saadatian Z, Hasani-Ranjbar S, Taslimi R, Bastami M, Amoli MM. Gholami M, et al. J Diabetes Metab Disord. 2019 Nov 26;18(2):675-692. doi: 10.1007/s40200-019-00459-2. eCollection 2019 Dec. J Diabetes Metab Disord. 2019. PMID: 31890692 Free PMC article. Review.
References
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. - PubMed
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. - PubMed
- Voorhoeve PM, Agami R. Classifying microRNAs in cancer: the good, the bad and the ugly. Biochim Biophys Acta. 2007;1775:274–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous