Human RAP1 inhibits non-homologous end joining at telomeres - PubMed (original) (raw)
Human RAP1 inhibits non-homologous end joining at telomeres
Jay Sarthy et al. EMBO J. 2009.
Abstract
Telomeres, the nucleoprotein structures at the ends of linear chromosomes, promote genome stability by distinguishing chromosome termini from DNA double-strand breaks (DSBs). Cells possess two principal pathways for DSB repair: homologous recombination and non-homologous end joining (NHEJ). Several studies have implicated TRF2 in the protection of telomeres from NHEJ, but the underlying mechanism remains poorly understood. Here, we show that TRF2 inhibits NHEJ, in part, by recruiting human RAP1 to telomeres. Heterologous targeting of hRAP1 to telomeric DNA was sufficient to bypass the need for TRF2 in protecting telomeric DNA from NHEJ in vitro. On expanding these studies in cells, we find that recruitment of hRAP1 to telomeres prevents chromosome fusions caused by the loss of TRF2/hRAP1 from chromosome ends despite activation of a DNA damage response. These results provide the first evidence that hRAP1 inhibits NHEJ at mammalian telomeres and identify hRAP1 as a mediator of genome stability.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
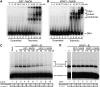
Figure 1
RAP1–TebDB binds telomeric DNA and inhibits NHEJ. (A) Electrophoretic mobility shift assay (EMSA) of double-stranded scrambled and telomeric DNA oligonucleotides incubated with indicated amounts of GST–TebDB. No protein was added in lanes 1 and 7. (B) EMSA of DNA substrates incubated with hRAP1–TebDB. (C) Inhibition of end joining by hRAP1–TebDB at telomeric DNA ends. Linear plasmid DNA containing twelve 5′-TTAGGG-3′ repeats at one end was incubated with GM00558 cell-free extract that was either mock depleted (lane 1) or immunodepleted (ID) of hRAP1 and TRF2 with anti-hRAP1 (lanes 2–10). GST–TebDB (lanes 3–6) and hRAP1–TebDB (lanes 7–10) were added at the indicated concentrations before incubation with DNA substrates. As each DNA substrate contains one telomeric (head) and one non-telomeric (tail) end, the presence of tail-to-tail fusions serves as an internal control for the presence of NHEJ activity in the extract (lane 1). End joining products were quantified by densitometry and were normalized to hRAP1-ID extract (lane 2). (D) Linear plasmid DNA containing twelve scrambled telomeric repeats (5′-TGAGTG-3′) at one end was incubated with GM00558 cell-free extract that was ID of hRAP1 and TRF2 with anti-hRAP1 (lanes 1–7). GST–TebDB (lanes 2–4) and hRAP1–TebDB (lanes 5–7) were added at the indicated concentrations before incubation with DNA substrates. End joining products were quantified by densitometry and were normalized to hRAP1-ID extract (lane 1). Gel was spliced to remove intervening lanes that were not pertinent to this experiment.
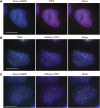
Figure 2
Telomeric localization of TebDB in human cells. (A) HeLa S3 cells transfected with Venus–TebDB (green) were stained with a mouse monoclonal antibody for TRF2 followed by an AlexaFluor 594-conjugated secondary antibody (red). Nuclei were visualized by counterstaining with DAPI (blue). Cells were subjected to nucleoplasmic extraction so that only chromatin-associated proteins remain within nuclei. (B) Cells expressing mCherry–TRF1 (red) were stained with a mouse monoclonal antibody for TRF2 and AlexaFluor 488 secondary antibody (green). (C) Visualization of Venus–TebDB (green) and mCherry–TRF1 (red) in co-transfected cells. All scale bars correspond to 10 μm.
Figure 3
Dominant-negative TRF2 (TRF2ΔBΔM) preferentially removes hRAP1 from telomeres. (A) Immunostaining of hRAP1 (AlexaFluor 488, green) and TRF2 (AlexaFluor 594, red) in HeLa S3 cells transfected with and selected for the presence of the vector controls. DNA was stained with DAPI (blue). (B) Visualization of TRF2 and hRAP1, as shown in (A), in cells expressing TRF2ΔBΔM. (C) Cells expressing TRF2ΔBΔM and Venus–TRF1 (green) were stained with anti-TRF2 (AlexaFluor 594, red) and DAPI (blue). (D) Cells, as shown in (C), were stained with anti-hRAP1 (AlexaFluor 594, red). All scale bars correspond to 10 μm.
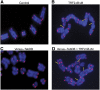
Figure 4
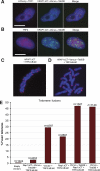
TebDB neither induces nor protects against NHEJ-dependent telomere fusions. Telomere FISH was carried out on metaphase spreads from cells transfected with and selected for (A) vector control, (B) TRF2ΔBΔM, (C) Venus-TebDB, (D) or Venus-TebDB and TRF2ΔBΔM. Telomeres were visualized with an AlexaFluor 543-labelled locked nucleic acid probe complementary to the G-rich strand (red). Chromosomes were counterstained with DAPI (blue). Representative chromosomes from the respective samples are shown. Some telomere–telomere fusions are highlighted with yellow arrows.
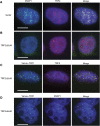
Figure 5
RAP1ΔCT–Venus–TebDB localizes to and protects telomeres in the absence of TRF2. (A) Co-localization of mCherry–TRF1 and hRAP1ΔCT–Venus–TebDB at telomeres in cells expressing TRF2ΔBΔM. (B) Cells expressing TRF2ΔBΔM and hRAP1ΔCT–Venus–TebDB were stained with anti-TRF2 and AlexaFluor 594-conjugated secondary antibody (red). Nucleoplasmic extraction was used to limit the visualization to chromatin-associated TRF2. hRAP1ΔCT–Venus–TebDB was visualized because of Venus fluorescence. Scale bars correspond to 10 μm. (C, D) Telomere FISH carried out on metaphase chromosomes transfected with and selected for expression of the indicated proteins. Telomeres were detected with an AlexaFluor 543-labelled locked nucleic acid probe that detects the G-rich strand (red). Chromosomes were stained with DAPI (blue). (E) Quantification of telomere fusions in metaphase spreads of cells transfected with and selected for the indicated constructs. Telomere fusions were quantified in images of metaphases from cells collected 72 h after transfection.
Figure 6
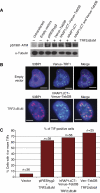
hRAP1 does not inhibit DNA damage signalling at telomeres. (A) ATM activation was analysed in extracts prepared from cells expressing the indicated proteins by immunoblotting with a phosphor-serine 1981-specific anti-ATM antibody. (B) Cells transfected with the indicated constructs were stained with an anti-53BP1 antibody. Telomeres were visualized by the green fluorescence of the Venus-containing fusions proteins. Scale bars represent 5 μm. (C) Quantification of telomere dysfunction-induced foci (TIFs) in cells expressing the indicated proteins. Cells with greater than four or more TIFs per cell were considered TIF positive.
Similar articles
- TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions.
Rai R, Chen Y, Lei M, Chang S. Rai R, et al. Nat Commun. 2016 Mar 4;7:10881. doi: 10.1038/ncomms10881. Nat Commun. 2016. PMID: 26941064 Free PMC article. - TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends.
Bombarde O, Boby C, Gomez D, Frit P, Giraud-Panis MJ, Gilson E, Salles B, Calsou P. Bombarde O, et al. EMBO J. 2010 May 5;29(9):1573-84. doi: 10.1038/emboj.2010.49. Epub 2010 Apr 20. EMBO J. 2010. PMID: 20407424 Free PMC article. - Human Rap1 interacts directly with telomeric DNA and regulates TRF2 localization at the telomere.
Arat NÖ, Griffith JD. Arat NÖ, et al. J Biol Chem. 2012 Dec 7;287(50):41583-94. doi: 10.1074/jbc.M112.415984. Epub 2012 Oct 20. J Biol Chem. 2012. PMID: 23086976 Free PMC article. - The Connection Between Cell Fate and Telomere.
Engin AB, Engin A. Engin AB, et al. Adv Exp Med Biol. 2021;1275:71-100. doi: 10.1007/978-3-030-49844-3_3. Adv Exp Med Biol. 2021. PMID: 33539012 Review. - No DDRama at chromosome ends: TRF2 takes centre stage.
Feuerhahn S, Chen LY, Luke B, Porro A. Feuerhahn S, et al. Trends Biochem Sci. 2015 May;40(5):275-85. doi: 10.1016/j.tibs.2015.03.003. Epub 2015 Apr 3. Trends Biochem Sci. 2015. PMID: 25845889 Review.
Cited by
- The Multifaceted Roles of Lamins in Lung Cancer and DNA Damage Response.
Janetzko J, Oeck S, Schramm A. Janetzko J, et al. Cancers (Basel). 2023 Nov 21;15(23):5501. doi: 10.3390/cancers15235501. Cancers (Basel). 2023. PMID: 38067205 Free PMC article. Review. - Genomic instability and telomere fusion of canine osteosarcoma cells.
Maeda J, Yurkon CR, Fujisawa H, Kaneko M, Genet SC, Roybal EJ, Rota GW, Saffer ER, Rose BJ, Hanneman WH, Thamm DH, Kato TA. Maeda J, et al. PLoS One. 2012;7(8):e43355. doi: 10.1371/journal.pone.0043355. Epub 2012 Aug 16. PLoS One. 2012. PMID: 22916246 Free PMC article. - A two-step mechanism for TRF2-mediated chromosome-end protection.
Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR 3rd, Denchi EL. Okamoto K, et al. Nature. 2013 Feb 28;494(7438):502-5. doi: 10.1038/nature11873. Epub 2013 Feb 6. Nature. 2013. PMID: 23389450 Free PMC article. - Telomeric protein TRF2 protects Holliday junctions with telomeric arms from displacement by the Werner syndrome helicase.
Nora GJ, Buncher NA, Opresko PL. Nora GJ, et al. Nucleic Acids Res. 2010 Jul;38(12):3984-98. doi: 10.1093/nar/gkq144. Epub 2010 Mar 9. Nucleic Acids Res. 2010. PMID: 20215438 Free PMC article. - Functional interaction between telomere protein TPP1 and telomerase.
Zaug AJ, Podell ER, Nandakumar J, Cech TR. Zaug AJ, et al. Genes Dev. 2010 Mar 15;24(6):613-22. doi: 10.1101/gad.1881810. Genes Dev. 2010. PMID: 20231318 Free PMC article.
References
- Bae NS, Baumann P (2007) A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell 26: 323–334 - PubMed
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 - PubMed
- Blue C, Marcand S, Gilson E (1997) Proteins that bind to double-stranded regions of telomeric DNA. Trends Cell Biol 7: 317–324 - PubMed
- Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous