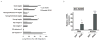SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas - PubMed (original) (raw)
. 2009 Nov;41(11):1238-42.
doi: 10.1038/ng.465. Epub 2009 Oct 4.
Hideo Watanabe, Craig H Mermel, Soyoung Yu, Sven Perner, Roel G Verhaak, So Young Kim, Leslie Wardwell, Pablo Tamayo, Irit Gat-Viks, Alex H Ramos, Michele S Woo, Barbara A Weir, Gad Getz, Rameen Beroukhim, Michael O'Kelly, Amit Dutt, Orit Rozenblatt-Rosen, Piotr Dziunycz, Justin Komisarof, Lucian R Chirieac, Christopher J Lafargue, Veit Scheble, Theresia Wilbertz, Changqing Ma, Shilpa Rao, Hiroshi Nakagawa, Douglas B Stairs, Lin Lin, Thomas J Giordano, Patrick Wagner, John D Minna, Adi F Gazdar, Chang Qi Zhu, Marcia S Brose, Ivan Cecconello, Ulysses Ribeiro Jr, Suely K Marie, Olav Dahl, Ramesh A Shivdasani, Ming-Sound Tsao, Mark A Rubin, Kwok K Wong, Aviv Regev, William C Hahn, David G Beer, Anil K Rustgi, Matthew Meyerson
- PMID: 19801978
- PMCID: PMC2783775
- DOI: 10.1038/ng.465
SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas
Adam J Bass et al. Nat Genet. 2009 Nov.
Abstract
Lineage-survival oncogenes are activated by somatic DNA alterations in cancers arising from the cell lineages in which these genes play a role in normal development. Here we show that a peak of genomic amplification on chromosome 3q26.33 found in squamous cell carcinomas (SCCs) of the lung and esophagus contains the transcription factor gene SOX2, which is mutated in hereditary human esophageal malformations, is necessary for normal esophageal squamous development, promotes differentiation and proliferation of basal tracheal cells and cooperates in induction of pluripotent stem cells. SOX2 expression is required for proliferation and anchorage-independent growth of lung and esophageal cell lines, as shown by RNA interference experiments. Furthermore, ectopic expression of SOX2 here cooperated with FOXE1 or FGFR2 to transform immortalized tracheobronchial epithelial cells. SOX2-driven tumors show expression of markers of both squamous differentiation and pluripotency. These characteristics identify SOX2 as a lineage-survival oncogene in lung and esophageal SCC.
Conflict of interest statement
Competing Interest Statement
Authors MM, WCH and RAS serve as consultants to Novartis. The authors know of no potential influence of this service upon the research presented in this manuscript.
Figures
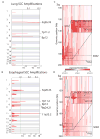
Figure 1. Recurrent genomic amplifications of 3q target SOX2 in lung and esophageal squamous cell carcinomas
A) Plots of recurrent high-level amplifications in 47 SCCs of the lung from GISTIC analysis of SNP array data. X-axis shows the G-score (top) and false discovery rate (q-value; bottom) for recurrent amplification across the genome with a green line demarcating an arbitrary FDR cut-off of 0.005. Labels on right denote the position of peaks of the most significantly altered regions. B) Depiction of GISTIC amplification peaks for 40 esophageal squamous cell carcinomas (29 primary tumors and 11 cell lines) C) Plot of copy-number data from chromosome 3q from lung SCC. Each sample is represented with a vertical line from centromere (top) to telomere (bottom). Areas of red indicate gain; blue indicates loss. The positions of SOX2 and TP63 are noted with horizontal lines. An inset box shows the 10-Mb region centered on SOX2 in greater detail in the 15 samples with highest SOX2 copy number. The grey lines depict the positions of the two nearest RefSeq genes to SOX2--ATP11B and _DNAJC19--_as well as PIK3CA. D) Plot of copy-number on chromosome 3q in esophageal SCC as described for panel C.
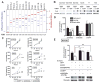
Figure 2. SOX2 knockdown via RNAi reduces anchorage-independent growth and proliferation of _SOX2_-overexpressing cell lines
A) RIGER analysis of shRNA against SOX2 and genes neighboring SOX2. Differential effects of each shRNA construct on proliferation of the four 3q26.33-amplified SCC cell lines was calculated by comparison of the effect of each shRNA construct in the SCC cell lines compared to the construct’s effect in two control lung adenocarcinoma cell lines. Blue lines represent differential proliferation scores for each shRNA construct. Negative enrichment scores represent reduced proliferation in the four SCC cell lines. Red lines represent the normalized enrichment score calculated for each gene based upon the proliferative effect of all shRNAs to that gene compared to effects of other shRNAs in this screen. False discovery rates (FDRs) for significant enrichment are listed below the graph; FDRs for SCC cell-specific reduced proliferation are shown in plain text and for control cell-specific reduced proliferation in italics. All results were normalized against the effects of control shRNAs (shGFP, shLacZ) in each cell line. B)Anti-Sox2 and control anti-vinculin immunoblots of lysates from established tumor cell lines stably expressing shRNA targeting SOX2 (shSOX2a or shSOX2b) or shRNA specific for green fluorescent protein (shGFP). HCC95 and NCI-H520 are lung SCC lines; TT and TE10 are esophageal SCC cell lines; and NCI-H1437 and NCI-H1355 are lung adenocarcinoma cell lines used as controls. C)Effect of _SOX2_-specific shRNA on viable cell numbers over time. Cells were measured at 24, 72 and 96 hours after plating and corrected to equalize 24-hr values. Mean cell viabilities (+/- standard deviations of cell plated in quadruplicate) are plotted as percentage of 24-hour measurement at 24, 72 and 96 hours after plating. (Note, due to low standard deviations of some measurements, error bars are not visible for all data points.) Significance levels are indicated with * marking p<0.05, ** for p<0.01 and *** for p<0.001. D)Soft agar colony formation for HCC95 and NCI-H520 and control NCI-H1437 cells expressing SOX2 shRNA is shown relative to shGFP (+/− standard deviation) with p-values marked as above. E) Soft agar colony formation for HCC95 cells engineered with ectopic expression of GFP, SOX2 or SOX2 R74P followed by infection with shSOX2b or shGFP. Data are shown relative to shGFP in HCC95-GFP cells (+/− standard deviation) with p-values marked as above. Immunoblots for Sox2 and vinculin are shown.
Figure 3. SOX2 can transform _FOXE1_- or _FGFR2_IIIb-expressing immortalized tracheobronchial epithelial cells
A) Soft agar colony formation for AALE tracheobronchial epithelial cells expressing either SOX2, FOXE1 or the combination of factors. Graph shows number of colonies (+/− standard deviation of experiment) with p-values labeled with asterisks as in Figure 2. Also pictured are representative soft-agar images and immunoblots showing expression of Sox2 and FoxE1. B) Soft agar colony formation data (+/− standard deviations), immunoblots and representative soft-agar images from co-transformation assays in AALE cells with SOX2 and FGFR2 IIIb and FGFR2 IIIc ectopic expression.
Figure 4. SOX2 induces expression of markers of both pluripotency and squamous differentiation
A) The percentages of lung SCC tumors showing over-expression of each of nine gene sets that are characteristically induced in ES cells are shown for samples with and without elevated SOX2 expression. Gene sets for which the FDR-corrected hypergeometric enrichment P-value for the differences in over-expression in cases with and without SOX2 over-expression are marked as in Figure 2. B) Quantitative RT-PCR for mRNA expression of squamous markers TP63 and KRT6A in NCI-H2009 cells with ectopic SOX2 compared to ectopic GFP with asterisks indicating p-values.
Similar articles
- SOX2 identified as a target gene for the amplification at 3q26 that is frequently detected in esophageal squamous cell carcinoma.
Gen Y, Yasui K, Zen Y, Zen K, Dohi O, Endo M, Tsuji K, Wakabayashi N, Itoh Y, Naito Y, Taniwaki M, Nakanuma Y, Okanoue T, Yoshikawa T. Gen Y, et al. Cancer Genet Cytogenet. 2010 Oct 15;202(2):82-93. doi: 10.1016/j.cancergencyto.2010.01.023. Cancer Genet Cytogenet. 2010. PMID: 20875870 - SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer.
Wilbertz T, Wagner P, Petersen K, Stiedl AC, Scheble VJ, Maier S, Reischl M, Mikut R, Altorki NK, Moch H, Fend F, Staebler A, Bass AJ, Meyerson M, Rubin MA, Soltermann A, Lengerke C, Perner S. Wilbertz T, et al. Mod Pathol. 2011 Jul;24(7):944-53. doi: 10.1038/modpathol.2011.49. Epub 2011 Apr 1. Mod Pathol. 2011. PMID: 21460799 - Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung.
Yuan P, Kadara H, Behrens C, Tang X, Woods D, Solis LM, Huang J, Spinola M, Dong W, Yin G, Fujimoto J, Kim E, Xie Y, Girard L, Moran C, Hong WK, Minna JD, Wistuba II. Yuan P, et al. PLoS One. 2010 Feb 9;5(2):e9112. doi: 10.1371/journal.pone.0009112. PLoS One. 2010. PMID: 20161759 Free PMC article. - Deciphering the cells of origin of squamous cell carcinomas.
Sánchez-Danés A, Blanpain C. Sánchez-Danés A, et al. Nat Rev Cancer. 2018 Sep;18(9):549-561. doi: 10.1038/s41568-018-0024-5. Nat Rev Cancer. 2018. PMID: 29849070 Free PMC article. Review. - The roles of the SOX2 protein in the development of esophagus and esophageal squamous cell carcinoma, and pharmacological target for therapy.
Zhang J, Wang Z, Zhao H, Wei Y, Zhou Y, Zhang S, Zhao J, Li X, Lin Y, Liu K. Zhang J, et al. Biomed Pharmacother. 2023 Jul;163:114764. doi: 10.1016/j.biopha.2023.114764. Epub 2023 Apr 24. Biomed Pharmacother. 2023. PMID: 37100016 Review.
Cited by
- Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers.
Cheung HW, Du J, Boehm JS, He F, Weir BA, Wang X, Butaney M, Sequist LV, Luo B, Engelman JA, Root DE, Meyerson M, Golub TR, Jänne PA, Hahn WC. Cheung HW, et al. Cancer Discov. 2011 Dec;1(7):608-25. doi: 10.1158/2159-8290.CD-11-0046. Epub 2011 Oct 17. Cancer Discov. 2011. PMID: 22586683 Free PMC article. - Epithelial cell plasticity: breaking boundaries and changing landscapes.
Tata A, Chow RD, Tata PR. Tata A, et al. EMBO Rep. 2021 Jul 5;22(7):e51921. doi: 10.15252/embr.202051921. Epub 2021 Jun 6. EMBO Rep. 2021. PMID: 34096150 Free PMC article. Review. - Increased Levels of Circulating and Tissue mRNAs of Oct-4, Sox-2, Bmi-1 and Nanog is ESCC Patients: Potential Tool for Minimally Invasive Cancer Diagnosis.
Bahl K, Saraya A, Sharma R. Bahl K, et al. Biomark Insights. 2012;7:27-37. doi: 10.4137/BMI.S8452. Epub 2012 Mar 26. Biomark Insights. 2012. PMID: 22493560 Free PMC article. - An optimal prognostic model based on gene expression for clear cell renal cell carcinoma.
Xu D, Dang W, Wang S, Hu B, Yin L, Guan B. Xu D, et al. Oncol Lett. 2020 Sep;20(3):2420-2434. doi: 10.3892/ol.2020.11780. Epub 2020 Jun 26. Oncol Lett. 2020. PMID: 32782559 Free PMC article. - Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development.
Lockwood WW, Wilson IM, Coe BP, Chari R, Pikor LA, Thu KL, Solis LM, Nunez MI, Behrens C, Yee J, English J, Murray N, Tsao MS, Minna JD, Gazdar AF, Wistuba II, MacAulay CE, Lam S, Lam WL. Lockwood WW, et al. PLoS One. 2012;7(5):e37775. doi: 10.1371/journal.pone.0037775. Epub 2012 May 21. PLoS One. 2012. PMID: 22629454 Free PMC article.
References
- Garraway L, Sellers W. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006;6:593–602. - PubMed
- Williamson KA, et al. Mutations in SOX2 cause anopthalmia esophageal-genital (AEG) syndrome. Hum Mol Genet. 2006;15:1413–1422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA090578-069005/CA/NCI NIH HHS/United States
- K08CA134931/CA/NCI NIH HHS/United States
- R33 CA128625/CA/NCI NIH HHS/United States
- K08 CA134931/CA/NCI NIH HHS/United States
- P01CA098101-05/CA/NCI NIH HHS/United States
- R01 CA109038/CA/NCI NIH HHS/United States
- R33 CA128625-01A1/CA/NCI NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
- R33CA128625/CA/NCI NIH HHS/United States
- P50 CA070907-029001/CA/NCI NIH HHS/United States
- R01 CA154365/CA/NCI NIH HHS/United States
- R01 CA071606/CA/NCI NIH HHS/United States
- P20 CA090578/CA/NCI NIH HHS/United States
- P50CA70907/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01CA109038/CA/NCI NIH HHS/United States
- T32 CA009172/CA/NCI NIH HHS/United States
- P01 CA098101-01A1/CA/NCI NIH HHS/United States
- P01 CA098101/CA/NCI NIH HHS/United States
- P50CA90578/CA/NCI NIH HHS/United States
- P50 CA090578/CA/NCI NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- R01CA071606-12/CA/NCI NIH HHS/United States
- K08 CA134931-01/CA/NCI NIH HHS/United States
- 5P01CA098101-05/CA/NCI NIH HHS/United States
- R01 CA071606-12/CA/NCI NIH HHS/United States
- DP1 OD003958/OD/NIH HHS/United States
- R01 CA109038-04/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous