Evolution in health and medicine Sackler colloquium: Somatic evolutionary genomics: mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration - PubMed (original) (raw)
Review
. 2010 Jan 26;107 Suppl 1(Suppl 1):1725-30.
doi: 10.1073/pnas.0909343106. Epub 2009 Sep 23.
Affiliations
- PMID: 19805033
- PMCID: PMC2868288
- DOI: 10.1073/pnas.0909343106
Review
Evolution in health and medicine Sackler colloquium: Somatic evolutionary genomics: mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration
Steven A Frank. Proc Natl Acad Sci U S A. 2010.
Abstract
Somatic mutations must happen often during development because of the large number of cell divisions to expand from a single-cell zygote to a full organism. A mutation in development carries forward to all descendant cells, causing genetic mosaicism. Widespread genetic mosaicism may influence diseases that derive from a few genetically altered cells, such as cancer. I show how to predict the expected amount of mosaicism and the variation in mosaicism between individuals. I then calculate the predicted risk of cancer derived from developmental mutations. The calculations show that a significant fraction of cancer in later life likely arises from developmental mutations in early life. In addition, much of the variation in the risk of cancer between individuals may arise from variation in the degree of genetic mosaicism set in early life. I also suggest that certain types of neurodegeneration, such as amyotrophic lateral sclerosis (ALS), may derive from a small focus of genetically altered cells. If so, then the risk of ALS would be influenced by developmental mutations and the consequent variation in genetic mosaicism. New technologies promise the ability to measure genetic mosaicism by sampling a large number of cellular genomes within an individual. The sampling of many genomes within an individual will eventually allow one to reconstruct the cell lineage history of genetic change in a single body. Somatic evolutionary genomics will follow from this technology, providing new insight into the origin and progression of disease with increasing age.
Conflict of interest statement
The author declares no conflict of interest.
Figures
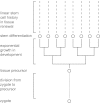
Fig. 1.
Lineage history of cells in renewing tissues. All cells trace their ancestry back to the zygote. Each tissue, or subset of tissue, derives from a precursor cell; _n_p rounds of cell division separate the precursor cell from the zygote. From a precursor cell, ne rounds of cell division lead to exponential clonal expansion until the descendants differentiate into the tissue-specific stem cells that seed the developing tissue. In a compartmental tissue, such as the intestine, lineage history of the renewing tissue follows an essentially linear path, in which each cellular history traces back through the same sequence of stem cell divisions (2, 21). At any point in time, a cell traces its history back through ns stem cell divisions to the ancestral stem cell in the tissue, and n = _n_p + ne + ns divisions back to the zygote. (Modified from figure 13.1 in ref. , based on the original in ref. .)
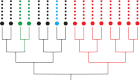
Fig. 2.
Mutational events in development occur at different stages in the exponential, branching phase of cellular expansion. In this example, the red mutation happens early, causing a significant fraction of somatic cells to carry the same mutation by descent. By contrast, the green mutation happens late in development, causing only a small fraction of somatic cells to carry the same mutation by descent. Many mutations will arise within the stem cells, each stem cell renewing only a very small fraction of all somatic cells. For example, the blue mutation is private to a single stem cell and will be confined to the small subset of somatic cells derived from that stem cell. Branching lines represent the developmental phase of cellular expansion, the large cells are stem cells, and the small cells in each line form a clone derived from their stem cell ancestor.
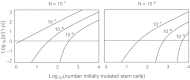
Fig. 3.
Number of initially mutated stem cells at the end of development. The N initial stem cells derive by exponential growth from a single precursor cell. Each plot shows the cumulative probability, p, for the number of mutated initial stem cells. By plotting log10(p/(1−p)), the zero line gives the median of the distribution. The number above each line is ue, the mutation probability per cell added to the population during exponential growth. (I used an actual value of 10−5.2 rather than 10−5 because of computational limitations.) For a single gene, the mutation probability per gene per cell division, _u_g, is probably >10−7. If there are at least L = 100 genes for which initial mutations can influence the progression to cancer, then ue = _Lu_g ≥ 10−5. Initial mutations may, for example, occur in DNA repair genes, causing an elevated rate of mutation at other loci. Calculations were made with algorithms in Zheng (38). (Modified from figure 13.3 in ref. , based on the original in ref. .)
Fig. 4.
The proportion of cancers that arise from cells mutated during development. These plots show calculations based on a specific four-stage model of colorectal cancer progression (22). The parameters of the progression model were estimated from incidence data. The values of u above each plot show the mutation rate per year in stem cells. Stem cells likely divide between 10 and 100 times per year, thus a mutation rate per year of at least 10−5 per locus seems reasonable. In each plot, the three curves sketch the heterogeneity between individuals in risk attributable to developmental mutations. The first quartile shows the proportion of cancers at each age for those individuals whose risk is in the lowest 25% of the population, in particular, those individuals who by chance have the fewest stem cells mutated during development. Similarly, the fourth quartile shows the risk for the highest 25% of the population with regard to developmental mutations. [Reproduced with permission from ref. (Copyright 2005, Elsevier).]
Similar articles
- Somatic mosaicism and disease.
Frank SA. Frank SA. Curr Biol. 2014 Jun 16;24(12):R577-R581. doi: 10.1016/j.cub.2014.05.021. Curr Biol. 2014. PMID: 24937287 Review. - Review: Somatic mutations in neurodegeneration.
Leija-Salazar M, Piette C, Proukakis C. Leija-Salazar M, et al. Neuropathol Appl Neurobiol. 2018 Apr;44(3):267-285. doi: 10.1111/nan.12465. Epub 2018 Feb 28. Neuropathol Appl Neurobiol. 2018. PMID: 29369391 Review. - Somatic mutations in neurodegeneration: An update.
Proukakis C. Proukakis C. Neurobiol Dis. 2020 Oct;144:105021. doi: 10.1016/j.nbd.2020.105021. Epub 2020 Jul 24. Neurobiol Dis. 2020. PMID: 32712267 Review. - Somatic mutations, genome mosaicism, cancer and aging.
Vijg J. Vijg J. Curr Opin Genet Dev. 2014 Jun;26:141-9. doi: 10.1016/j.gde.2014.04.002. Epub 2014 Oct 2. Curr Opin Genet Dev. 2014. PMID: 25282114 Free PMC article. Review. - Somatic mutations in neurons during aging and neurodegeneration.
Verheijen BM, Vermulst M, van Leeuwen FW. Verheijen BM, et al. Acta Neuropathol. 2018 Jun;135(6):811-826. doi: 10.1007/s00401-018-1850-y. Epub 2018 Apr 28. Acta Neuropathol. 2018. PMID: 29705908 Free PMC article. Review.
Cited by
- Onco-Breastomics: An Eco-Evo-Devo Holistic Approach.
Neagu AN, Whitham D, Bruno P, Arshad A, Seymour L, Morrissiey H, Hukovic AI, Darie CC. Neagu AN, et al. Int J Mol Sci. 2024 Jan 28;25(3):1628. doi: 10.3390/ijms25031628. Int J Mol Sci. 2024. PMID: 38338903 Free PMC article. Review. - A microglia clonal inflammatory disorder in Alzheimer's Disease.
Vicario R, Fragkogianni S, Weber L, Lazarov T, Hu Y, Hayashi SY, Craddock BP, Socci ND, Alberdi A, Baako A, Ay O, Ogishi M, Lopez-Rodrigo E, Kappagantula R, Viale A, Iacobuzio-Donahue CA, Zhou T, Ransohoff RM, Chesworth R; Netherlands Brain Bank; Abdel-Wahab O, Boisson B, Elemento O, Casanova JL, Miller WT, Geissmann F. Vicario R, et al. bioRxiv [Preprint]. 2024 Aug 3:2024.01.25.577216. doi: 10.1101/2024.01.25.577216. bioRxiv. 2024. PMID: 38328106 Free PMC article. Preprint. - Cancer Prevalence Across Vertebrates.
Compton ZT, Harris V, Mellon W, Rupp S, Mallo D, Kapsetaki SE, Wilmot M, Kennington R, Noble K, Baciu C, Ramirez L, Peraza A, Martins B, Sudhakar S, Aksoy S, Furukawa G, Vincze O, Giraudeau M, Duke EG, Spiro S, Flach E, Davidson H, Zehnder A, Graham TA, Troan B, Harrison TM, Tollis M, Schiffman JD, Aktipis A, Abegglen LM, Maley CC, Boddy AM. Compton ZT, et al. Res Sq [Preprint]. 2023 Jul 6:rs.3.rs-3117313. doi: 10.21203/rs.3.rs-3117313/v1. Res Sq. 2023. PMID: 37461608 Free PMC article. Updated. Preprint. - Opinion: more mouse models and more translation needed for ALS.
Fisher EMC, Greensmith L, Malaspina A, Fratta P, Hanna MG, Schiavo G, Isaacs AM, Orrell RW, Cunningham TJ, Arozena AA. Fisher EMC, et al. Mol Neurodegener. 2023 May 4;18(1):30. doi: 10.1186/s13024-023-00619-2. Mol Neurodegener. 2023. PMID: 37143081 Free PMC article. Review. - The origins and functional effects of postzygotic mutations throughout the human life span.
Rockweiler NB, Ramu A, Nagirnaja L, Wong WH, Noordam MJ, Drubin CW, Huang N, Miller B, Todres EZ, Vigh-Conrad KA, Zito A, Small KS, Ardlie KG, Cohen BA, Conrad DF. Rockweiler NB, et al. Science. 2023 Apr 14;380(6641):eabn7113. doi: 10.1126/science.abn7113. Epub 2023 Apr 14. Science. 2023. PMID: 37053313 Free PMC article.
References
- Frank SA, Nowak MA. Developmental predisposition to cancer. Nature. 2003;422:494. - PubMed
- Frank SA. Dynamics of Cancer: Incidence, Inheritance, and Evolution. Princeton: Princeton Univ Press; 2007. - PubMed
- Araten DJ, et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–8117. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous