One billion years of p53/p63/p73 evolution - PubMed (original) (raw)
Comment
One billion years of p53/p63/p73 evolution
Vladimir A Belyi et al. Proc Natl Acad Sci U S A. 2009.
No abstract available
Conflict of interest statement
The authors declare no conflict of interest.
Figures
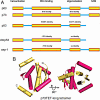
Fig. 1.
The domain structure of the p53 family proteins. (A) The domain organization of human p53, p63, and p73 along with the p63/p73 ancestor proteins from Drosophila melanogaster and Caenorhabditis elegans. The percentage of identity of the amino acids and their position in the domain are given for both the DNA-binding domain and the oligomeric domain. (B) The structure of the oligomeric domain of p73 from ref. . The H-2 helix stabilizes this p73 tetramer and is absent from the p53 tetramer. p63 and p73 are more closely related to each other than to p53 based on the primary, secondary, tertiary, and quaternary structures of both the DNA-binding domains and the oligomerization domains.
Comment on
- Structural evolution of p53, p63, and p73: implication for heterotetramer formation.
Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR. Joerger AC, et al. Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17705-10. doi: 10.1073/pnas.0905867106. Epub 2009 Oct 7. Proc Natl Acad Sci U S A. 2009. PMID: 19815500 Free PMC article.
Similar articles
- Structural evolution of p53, p63, and p73: implication for heterotetramer formation.
Joerger AC, Rajagopalan S, Natan E, Veprintsev DB, Robinson CV, Fersht AR. Joerger AC, et al. Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17705-10. doi: 10.1073/pnas.0905867106. Epub 2009 Oct 7. Proc Natl Acad Sci U S A. 2009. PMID: 19815500 Free PMC article. - The functional domains in p53 family proteins exhibit both common and distinct properties.
Harms KL, Chen X. Harms KL, et al. Cell Death Differ. 2006 Jun;13(6):890-7. doi: 10.1038/sj.cdd.4401904. Cell Death Differ. 2006. PMID: 16543939 Review. No abstract available. - Therapeutic prospects for p73 and p63: rising from the shadow of p53.
Vilgelm A, El-Rifai W, Zaika A. Vilgelm A, et al. Drug Resist Updat. 2008 Aug-Oct;11(4-5):152-63. doi: 10.1016/j.drup.2008.08.001. Epub 2008 Sep 17. Drug Resist Updat. 2008. PMID: 18801697 Free PMC article. Review. - Tracing the protectors path from the germ line to the genome.
Coutandin D, Ou HD, Löhr F, Dötsch V. Coutandin D, et al. Proc Natl Acad Sci U S A. 2010 Aug 31;107(35):15318-25. doi: 10.1073/pnas.1001069107. Epub 2010 Aug 9. Proc Natl Acad Sci U S A. 2010. PMID: 20696896 Free PMC article. - p63 and p73, the ancestors of p53.
Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. Dötsch V, et al. Cold Spring Harb Perspect Biol. 2010 Sep;2(9):a004887. doi: 10.1101/cshperspect.a004887. Epub 2010 May 19. Cold Spring Harb Perspect Biol. 2010. PMID: 20484388 Free PMC article. Review.
Cited by
- Do Mutations Turn p53 into an Oncogene?
Pitolli C, Wang Y, Mancini M, Shi Y, Melino G, Amelio I. Pitolli C, et al. Int J Mol Sci. 2019 Dec 11;20(24):6241. doi: 10.3390/ijms20246241. Int J Mol Sci. 2019. PMID: 31835684 Free PMC article. Review. - The structure of the FYR domain of transforming growth factor beta regulator 1.
García-Alai MM, Allen MD, Joerger AC, Bycroft M. García-Alai MM, et al. Protein Sci. 2010 Jul;19(7):1432-8. doi: 10.1002/pro.404. Protein Sci. 2010. PMID: 20506279 Free PMC article. - Dipeptide analysis of p53 mutations and evolution of p53 family proteins.
Huang Q, Yu L, Levine AJ, Nussinov R, Ma B. Huang Q, et al. Biochim Biophys Acta. 2014 Jan;1844(1 Pt B):198-206. doi: 10.1016/j.bbapap.2013.04.002. Epub 2013 Apr 10. Biochim Biophys Acta. 2014. PMID: 23583620 Free PMC article. - An insight into cancer palaeobiology: does the Mesozoic neoplasm support tissue organization field theory of tumorigenesis?
Surmik D, Słowiak-Morkovina J, Szczygielski T, Kamaszewski M, Kalita S, Teschner EM, Dróżdż D, Duda P, Rothschild BM, Konietzko-Meier D. Surmik D, et al. BMC Ecol Evol. 2022 Dec 13;22(1):143. doi: 10.1186/s12862-022-02098-3. BMC Ecol Evol. 2022. PMID: 36513967 Free PMC article. - Molecular dynamics of the full-length p53 monomer.
Chillemi G, Davidovich P, D'Abramo M, Mametnabiev T, Garabadzhiu AV, Desideri A, Melino G. Chillemi G, et al. Cell Cycle. 2013 Sep 15;12(18):3098-108. doi: 10.4161/cc.26162. Epub 2013 Sep 5. Cell Cycle. 2013. PMID: 23974096 Free PMC article.
References
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. - PubMed
- Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. - PubMed
- Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. - PubMed
- Celli J, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous