A neural circuit mechanism integrating motivational state with memory expression in Drosophila - PubMed (original) (raw)
A neural circuit mechanism integrating motivational state with memory expression in Drosophila
Michael J Krashes et al. Cell. 2009.
Abstract
Behavioral expression of food-associated memory in fruit flies is constrained by satiety and promoted by hunger, suggesting an influence of motivational state. Here, we identify a neural mechanism that integrates the internal state of hunger and appetitive memory. We show that stimulation of neurons that express neuropeptide F (dNPF), an ortholog of mammalian NPY, mimics food deprivation and promotes memory performance in satiated flies. Robust appetitive memory performance requires the dNPF receptor in six dopaminergic neurons that innervate a distinct region of the mushroom bodies. Blocking these dopaminergic neurons releases memory performance in satiated flies, whereas stimulation suppresses memory performance in hungry flies. Therefore, dNPF and dopamine provide a motivational switch in the mushroom body that controls the output of appetitive memory.
Figures
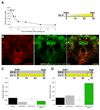
Figure 1. Stimulating dNPF neurons promotes appetitive memory expression in satiated flies
A. Feeding after training suppresses appetitive memory performance. Double asterisk, significant difference (P<0.007); single asterisk (P<0.03) from all other groups. Temperature shift protocols are shown. White bar represents fly storage in empty vials, while yellow bar indicates flies stored with food -figure format used throughout this study. B. dNPF is expressed in neurons that innervate the dorsal and lateral protocerebrum, the SOG and the CC. Immunostaining with anti-dNPF antibody (red), partially overlaps (yellow, merge) with _dNPF_-GAL4 driven CD8::GFP (green). dNPF stained cells in the SOG are not labeled by _dNPF_-GAL4. The dNPF antibody only labels the upper layer of the fan-shaped body, consistent with processes in the ellipsoid body and lower fan-shaped body being post-synaptic regions. Scale bar represents 20µm. C. Feeding flies after training suppresses memory performance. All flies were food-deprived, trained, fed and tested at 23°C. D. Stimulating dNPF neurons before testing produces memory performance in fed flies. All flies were food-deprived, trained, and fed for 150min at 23°C. All flies were then transferred to 31°C for 30min and tested for appetitive memory. Asterisk denotes significant difference (P<0.05, ANOVA) from unmarked groups. Data are mean ± standard error of the mean (SEM).
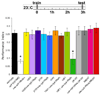
Figure 2. Disruption of npfr1 expression impairs appetitive memory in food-deprived flies
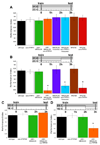
A. Food-deprived _npfr1_[c01896] flies and those expressing uas-_npfr1_RNAi with c061 have impaired 3hr appetitive memory whereas those expressing uas-_npfr1_RNAi with c005, 210Y, 104Y, OK107 or MB247 are normal. Asterisk denotes significant difference (P<0.01, ANOVA) from other unmarked groups. Data are mean ± SEM.
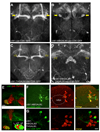
Figure 3. c061 labels six DA neurons that innervate the MB
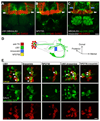
A. Projection view of a c061;uas-CD8::GFP brain. Filled yellow arrows mark the heel region of the MB resembling that in _TH_-GAL4 labeled brains (Figure S4A). B. Combining MBGAL80 with c061;uas-CD8::GFP eliminates MB expression and reveals neurons that innervate the heel of the MB (filled yellow arrows). Also see Movie S1 and Movie S2. C. Combining _TH_GAL80 with c061;uas-CD8::GFP eliminates expression in the neurons innervating the region between the MB lobes and MB heel (hollow arrows) but leaves expression elsewhere intact. D. Projection view of a c061;MBGAL80/THGAL80;uas-CD8::GFP brain. _TH_GAL80 removes expression from the neurons innervating the dorsal protocerebrum and MB heel (hollow arrows) and leaves expression in the fan-shaped body and elsewhere intact. Scale bar represents 20 µm. E. Higher magnification single confocal section views of the MB heel and peduncle region in a c061; uas-CD8::GFP brain. Top panel shows innervation of the MB heel. Bottom panel, innervation in the base of the peduncle. Inset, section through the peduncle showing zones occupied by the αβ,α´β´ and γ MB neurons. The MB is co-labeled with MB-DsRED. F. Confocal section through a c061;MBGAL80;uas-CD8::GFP brain at the level of the MB calyx (outlined). GFP (green) labels 3 cell bodies at the side of the calyx and 5 more lateral cell bodies. Counter staining with anti-TH antibody (red) labels 12 cell bodies in the PPL1 cluster and 3 of them overlap (merge, yellow) with c061;MBGAL80 driven GFP. Scale bar represents 10 µm. G. Confocal section through a PPL1 cluster in a c061;MBGAL80/_TH_GAL80;uas-CD8::GFP brain. GFP (green) labels 5 cell bodies and none overlap with anti-TH staining. Scale bar represents 10 µm.
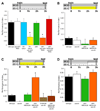
Figure 4. c061 DA neurons regulate appetitive memory performance
Temperature shift protocols are shown above each graph. A. Expressing uas-npfr1RNAi in all DA neurons with _TH_-GAL4 or in a subset of DA neurons with c061 impairs 3hr appetitive memory in hungry flies. Expressing uas-npfr1RNAi in c061 neurons except the DA neurons (c061;_TH_GAL80;uas-npfr1RNAi) does not affect appetitive memory. B. Feeding flies after training suppresses 3hr memory. All genotypes were food-deprived, trained, fed and tested at the permissive temperature of 23°C. C. Blocking synaptic output from c061;MBGAL80 neurons before testing reveals memory performance in fed flies. Removing uas-_shi_ts1 expression from the DA neurons reverses the memory promoting effect (c061;MBGAL80/_TH_GAL80;uas-_shi_ts1 flies). All genotypes were food-deprived, trained and stored in food vials for 120min at 23°C. Flies were then shifted to 31°C for 60min and tested for memory. D. Blocking output from c061;MBGAL80 neurons does not enhance performance in food-deprived flies. All genotypes were food-deprived, trained and stored in empty vials for 120min at 23°C. Flies were then shifted to 31°C for 60min before flies were tested for appetitive memory at 31°C. Asterisks denote significant difference (P<0.05, ANOVA) from other unmarked groups. Data are mean ± SEM.
Figure 5. The c061 DA neurons are MB-MP neurons
A. Projection view of a c061;MBGAL80;uas-CD8::GFP brain showing MB-MP neurons (white arrows) and neurons in the subesophageal ganglion (also see Movie S1 and Movie S2). B. Projection view of a NP2758;uas-CD8::GFP brain showing MB-MP neurons (white arrows) and neurons in the subesophageal ganglion (also see Movie S3 and Movie S4). C. Projection view of a MBGAL80; krasavietz/uas-CD8::GFP brain showing MB-MP neurons (white arrows). DA neurons innervating the MB α-stalk (blue arrows, also see Figure S4C), and neurons in the fan-shaped body and local neurons in the antennal lobe are also labeled (also see Movie S5 and Movie S6). The MB is labeled with MB-DsRED. D. Cartoon illustrating the gross structure of MB-MP neurons and the expression pattern of each GAL4. The MB is shown as an outline. DA neuron cell bodies (red, anti-TH) of a single PPL1 cluster are shown with the labeling of each GAL4 overlayed. The cell body organization is not stereotyped and it is difficult to distinguish the projections of each MB-MP neuron. No order or detail is inferred here. At least one MB-MP neuron sends a contralateral projection to the other MB (green arrow head). E. krasavietz and NP2758 label a subset of c061 MB-MP neurons. Each column shows the separate and merged channels from confocal images of a PPL1 cluster in brains co-labeled with GAL4 driven GFP (green) and anti-TH (red). Double-labeled neurons are marked with an arrow in the merged images. The quantification of neurons is shown in Figure S4B. Scale bar represents 20 µm (A, B, C) or 10 µm (E).
Figure 6. MB-MP stimulation suppresses appetitive memory expression in hungry flies
Temperature shift protocols are shown above each graph. A. The permissive 23°C does not disrupt 3hr appetitive memory. All genotypes were starved, trained, stored for 3hr in empty vials and tested for memory at 23°C. B. Stimulating 6, 4 or 2 MB-MP neurons with uas-dTrpA1 before and during testing attenuates memory performance in starved flies. All genotypes were food-deprived, trained and stored in empty vials for 120min at 23°C and were then shifted to 31°C for 60min before and during testing. C. The permissive temperature of 23°C does not disrupt 3hr appetitive memory. All genotypes were starved, trained, stored in empty vials for 3 hr and tested for appetitive memory at 23°C. D. Stimulating 6 MB-MP neurons with uas-TRPM8 before and during testing attenuates memory performance in starved flies. All genotypes were food-deprived, trained and stored in empty food vials for 120min at 23°C and were then shifted to 16°C for 60min before and during testing. Asterisk denotes significant difference (P<0.05, ANOVA) from other unmarked groups. Data are mean ± SEM.
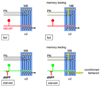
Figure 7. Model for the role of MB-MP neurons
Left panels illustrate the state of the inhibitory control exerted upon the MB in the fed state (top) and starved state (bottom). When fed flies are exposed to the conditioned odor during memory testing (right panels) the appropriate projection neurons and MB neurons are activated (yellow). However, the signal only propagates beyond the MB neurons in hungry flies when the MB-MP neuron ‘gate’ is open. Red lines denote inhibition and green lines relief from inhibition. See discussion for more detail.
Comment in
- New routes for memory retrieval and reinforcement.
Dean T, Sehgal A. Dean T, et al. Cell. 2009 Oct 16;139(2):225-7. doi: 10.1016/j.cell.2009.09.031. Cell. 2009. PMID: 19837025
Similar articles
- Appetitive Memory with Survival Benefit Is Robust Across Aging in Drosophila.
Tonoki A, Ogasawara M, Yu Z, Itoh M. Tonoki A, et al. J Neurosci. 2020 Mar 11;40(11):2296-2304. doi: 10.1523/JNEUROSCI.2045-19.2020. Epub 2020 Jan 28. J Neurosci. 2020. PMID: 31992587 Free PMC article. - Drosophila mushroom bodies integrate hunger and satiety signals to control innate food-seeking behavior.
Tsao CH, Chen CC, Lin CH, Yang HY, Lin S. Tsao CH, et al. Elife. 2018 Mar 16;7:e35264. doi: 10.7554/eLife.35264. Elife. 2018. PMID: 29547121 Free PMC article. - Aversive Learning and Appetitive Motivation Toggle Feed-Forward Inhibition in the Drosophila Mushroom Body.
Perisse E, Owald D, Barnstedt O, Talbot CB, Huetteroth W, Waddell S. Perisse E, et al. Neuron. 2016 Jun 1;90(5):1086-99. doi: 10.1016/j.neuron.2016.04.034. Epub 2016 May 19. Neuron. 2016. PMID: 27210550 Free PMC article. - Dopamine reveals neural circuit mechanisms of fly memory.
Waddell S. Waddell S. Trends Neurosci. 2010 Oct;33(10):457-64. doi: 10.1016/j.tins.2010.07.001. Epub 2010 Aug 10. Trends Neurosci. 2010. PMID: 20701984 Free PMC article. Review. - Gustatory processing and taste memory in Drosophila.
Masek P, Keene AC. Masek P, et al. J Neurogenet. 2016 Jun;30(2):112-21. doi: 10.1080/01677063.2016.1185104. J Neurogenet. 2016. PMID: 27328844 Free PMC article. Review.
Cited by
- The Neural Correlations of Olfactory Associative Reward Memories in Drosophila.
Lin YC, Wu T, Wu CL. Lin YC, et al. Cells. 2024 Oct 17;13(20):1716. doi: 10.3390/cells13201716. Cells. 2024. PMID: 39451234 Free PMC article. Review. - Independent operations of appetitive and aversive conditioning systems lead to simultaneous production of conflicting memories in an insect.
Rahman S, Terao K, Hashimoto K, Mizunami M. Rahman S, et al. Proc Biol Sci. 2024 Sep;291(2031):20241273. doi: 10.1098/rspb.2024.1273. Epub 2024 Sep 25. Proc Biol Sci. 2024. PMID: 39317316 - A LAT1-Like Amino Acid Transporter Regulates Neuronal Activity in the Drosophila Mushroom Bodies.
Delescluse J, Simonnet MM, Ziegler AB, Piffaretti K, Alves G, Grosjean Y, Manière G. Delescluse J, et al. Cells. 2024 Aug 13;13(16):1340. doi: 10.3390/cells13161340. Cells. 2024. PMID: 39195231 Free PMC article. - An integrative sensor of body states: how the mushroom body modulates behavior depending on physiological context.
Suárez-Grimalt R, Grunwald Kadow IC, Scheunemann L. Suárez-Grimalt R, et al. Learn Mem. 2024 Jun 14;31(5):a053918. doi: 10.1101/lm.053918.124. Print 2024 May. Learn Mem. 2024. PMID: 38876486 Free PMC article. Review. - Roles of feedback and feed-forward networks of dopamine subsystems: insights from Drosophila studies.
Davidson AM, Hige T. Davidson AM, et al. Learn Mem. 2024 Jun 11;31(5):a053807. doi: 10.1101/lm.053807.123. Print 2024 May. Learn Mem. 2024. PMID: 38862171 Free PMC article. Review.
References
- Andretic R, van Swinderen B, Greenspan RJ. Dopaminergic modulation of arousal in Drosophila. Curr Biol. 2005;15:1165–1175. - PubMed
- Arquier N, Geminard C, Bourouis M, Jarretou G, Honegger B, Paix A, Leopold P. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 2008;7:333–338. - PubMed
- Bewick GA, Gardiner JV, Dhillo WS, Kent AS, White NE, Webster Z, Ghatei MA, Bloom SR. Post-embryonic ablation of AgRP neurons in mice leads to a lean, hypophagic phenotype. FASEB J. 2005;19:1680–1682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MH081982/MH/NIMH NIH HHS/United States
- R01 MH081982/MH/NIMH NIH HHS/United States
- DA024499/DA/NIDA NIH HHS/United States
- F31 DA024499/DA/NIDA NIH HHS/United States
- R01 MH081982-01A1/MH/NIMH NIH HHS/United States
- MH09883/MH/NIMH NIH HHS/United States
- R01 MH069883-06A2/MH/NIMH NIH HHS/United States
- R01 MH069883/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous