Runx proteins regulate Foxp3 expression - PubMed (original) (raw)
. 2009 Oct 26;206(11):2329-37.
doi: 10.1084/jem.20090226. Epub 2009 Oct 19.
Luca Mazzarella, Maarten Hoogenkamp, Arnulf Hertweck, Bradley S Cobb, Stephan Sauer, Suzana Hadjur, Marion Leleu, Yoshinori Naoe, Janice C Telfer, Constanze Bonifer, Ichiro Taniuchi, Amanda G Fisher, Matthias Merkenschlager
Affiliations
- PMID: 19841090
- PMCID: PMC2768863
- DOI: 10.1084/jem.20090226
Runx proteins regulate Foxp3 expression
Ludovica Bruno et al. J Exp Med. 2009.
Abstract
Runx proteins are essential for hematopoiesis and play an important role in T cell development by regulating key target genes, such as CD4 and CD8 as well as lymphokine genes, during the specialization of naive CD4 T cells into distinct T helper subsets. In regulatory T (T reg) cells, the signature transcription factor Foxp3 interacts with and modulates the function of several other DNA binding proteins, including Runx family members, at the protein level. We show that Runx proteins also regulate the initiation and the maintenance of Foxp3 gene expression in CD4 T cells. Full-length Runx promoted the de novo expression of Foxp3 during inducible T reg cell differentiation, whereas the isolated dominant-negative Runt DNA binding domain antagonized de novo Foxp3 expression. Foxp3 expression in natural T reg cells remained dependent on Runx proteins and correlated with the binding of Runx/core-binding factor beta to regulatory elements within the Foxp3 locus. Our data show that Runx and Foxp3 are components of a feed-forward loop in which Runx proteins contribute to the expression of Foxp3 and cooperate with Foxp3 proteins to regulate the expression of downstream target genes.
Figures
Figure 1.
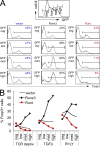
Foxp3 induction is promoted by full-length Runx3 and antagonized by the expression of dominant-negative Runt domain proteins. (A) Naive CD4 T cells were activated for 18 h with anti-CD3/anti-CD28 beads and retrovirally transduced with Runx3-IRES-GFP (Runx3), Runt-IRES-GFP (Runt), or control-IRES-GFP (vector). After 24 h, 0.3 ng/ml TGF-β was added and Foxp3 expression was assessed 48 h later by intracellular staining after gating on the level of GFP expression (negative [neg], low, medium [med], or high). Data are representative of four independent experiments. (B) Naive CD4 T cells were activated and retrovirally transduced as in A. After 24 h, the anti-CD3/anti-CD28 beads were removed (TCR depriv.) and TGF-β or rapamycin and LY294002 (R+LY) were added as indicated. Foxp3 expression was assessed 48 h later by intracellular staining after gating on the level of GFP expression (negative, low, medium, or high). Data are representative of four independent experiments.
Figure 2.
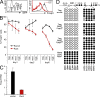
Runx proteins control the maintenance of Foxp3 expression in established T reg cells. (A) CD4+ CD25+ LN T cells were sorted, activated for 18 h with anti-CD3/anti-CD28 beads in the presence of IL-2, and retrovirally transduced. Histograms show GFP expression (left) and Foxp3 expression (right) in GFPhigh cells 4 d after transduction with vector-IRES-GFP (black line, right histogram) or Runt-IRES-GFP (red line, right histogram). Data are representative of five independent experiments. (B) CD4+ CD25+ LN T cells were sorted, activated, and retrovirally transduced with Runx3-IRES-GFP (black) or Runt-IRES-GFP (red), and Foxp3 expression was assessed on the indicated days by intracellular staining and gating on the level of GFP (negative, low, medium, or high as defined in A). Data are mean ± SE of two independent experiments. (C) CD4+ CD25+ LN T cells were sorted, activated, and retrovirally transduced with IRES-GFP (vector) or Runt-IRES-GFP (Runt). 5 d later, GFPhigh cells were sorted and analyzed for Foxp3 RNA expression by real time RT-PCR. Data are mean ± SD of five independent experiments. (D) Runt-transduced T reg cells with down-regulated Foxp3 expression have a demethylated Foxp3 DMR, which is consistent with a history of Foxp3 expression. CD4+ CD25+ LN T cells were sorted, activated, and transduced with Runt-IRES-GFP (Runt). 5 d later, GFPhigh Foxp3high and Foxp3low cells were sorted and analyzed for the methylation status of the indicated regions in the Foxp3 locus by bisulfite sequencing. A schematic of the locus with coding (filled rectangles) and noncoding exons (open rectangles) is shown on top. Each chain of circles represents one sequenced allele with methylated (filled circles) and unmethylated CpG dinucleotides (open circles). The positions of CpG dinucleotides are indicated (Floess et al., 2007).
Figure 3.
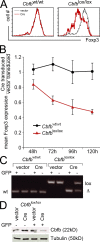
Genetic evidence for a role of Runx proteins in the maintenance of Foxp3 expression in nT reg cells. (A) CD4+ CD25+ LN T cells were sorted from control _Cbfb_wt/wt and _Cbfb_lox/lox mice, activated for 18 h with anti-CD3/anti-CD28 beads in the presence of IL-2, and retrovirally transduced with vector-IRES-GFP (vector) or Cre-IRES-GFP (Cre). 4 d later, Foxp3 expression had declined selectively in _Cbfb_lox/lox cells transduced with Cre-IRES-GFP (red). Foxp3 expression remained unaffected in vector-transduced _Cbfb_lox/lox cells (black) and in _Cbfb_wt/wt cells transduced with either vector-IRES-GFP (black) or Cre-IRES-GFP (red). Data are representative of three independent experiments. (B) Time course of Foxp3 expression by GFP+ Cre-IRES-GFP–transduced relative to vector-IRES-GFP–transduced _Cbfb_lox/lox nT reg cells (red) and _Cbfb_wt/wt nT reg cells (black; mean ± SD of three independent experiments). (C) GFP-positive and -negative nT reg cells were sorted 4 d after transduction with vector-IRES-GFP (vector) or Cre-IRES-GFP (Cre), and Cbfb loci were analyzed by genomic PCR. The position of bands corresponding to the wild-type (wt), floxed (lox), and deleted (Δ) locus are indicated. One of three independent experiments is shown. (D) GFP-positive and -negative _Cbfb_lox/lox cells were sorted 4 d after transduction with vector-IRES-GFP (vector) or Cre-IRES-GFP (Cre), and Cbfb protein expression was analyzed by Western blotting. Tubulin was used as a loading control. One of three independent experiments is shown.
Figure 4.
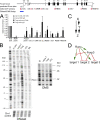
Runx and Foxp3 as components of a feed-forward loop. (A) Cbfb ChIP in naive CD4 T cells (gray bars) and ex vivo–isolated T reg cells (black bars). The Tcrb enhancer (enh.) and Myf5 promoter (prom.) are used as positive and negative controls, respectively. Predicted RBSs are indicated, and sites 2, 3, 4, and 5 are conserved between mouse and human. Regions within the Foxp3 locus examined for Cbfb binding are indicated in red. IgG is used instead of anti-Cbfb as a ChIP control. The region examined by footprint analysis is in blue. PCR signals are normalized to input and shown relative to the positive control site Tcrb enh. (mean ± SD; n = 3 independent experiments). (B) DNaseI (left) and DMS footprinting (right) of the Foxp3 promoter region. G, G sequencing reaction; in vitro, naked DNA; naive T, naive CD4 T cells; T reg, ex vivo–isolated T reg cells; 3T3, NIH 3T3 cells; Mφ, macrophages. Base positions are shown relative to the Foxp3 transcription start site. Note that T reg cells show increased DNaseI accessibility of the Foxp3 promoter (stronger bands in the T reg sample) but not of a control region on chromosome 1. Vertical lines indicate local DNaseI sensitivity in T reg cells. Open circles indicate DMS-protected (weaker) bands in T reg cells, which may indicate the position of DNA binding proteins. A predicted Runx site is indicated. Data are representative of an extensive analysis of both forward and reverse strands with primers for the proximal and distal Foxp3 promoter region using independent template preparations for DNaseI and DMS treatment. (C) Scheme of a feed-forward loop where X regulates Y and X and Y together regulate Z. (D) Model describing a relationship between Runx and Foxp3 where Runx acts on Foxp3 expression and Foxp3 either blocks Runx activity, converts Runx from an activator into a repressor, or cooperates with Runx to activate or repress downstream target genes.
Similar articles
- Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells.
Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, Rückert B, Meiler F, Akdis M, Littman DR, Akdis CA. Klunker S, et al. J Exp Med. 2009 Nov 23;206(12):2701-15. doi: 10.1084/jem.20090596. Epub 2009 Nov 16. J Exp Med. 2009. PMID: 19917773 Free PMC article. - Induction of CD4+CD25+Foxp3+ regulatory T cells by mesenchymal stem cells is associated with RUNX complex factors.
Khosravi M, Bidmeshkipour A, Moravej A, Hojjat-Assari S, Naserian S, Karimi MH. Khosravi M, et al. Immunol Res. 2018 Feb;66(1):207-218. doi: 10.1007/s12026-017-8973-4. Immunol Res. 2018. PMID: 29143918 - Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells.
Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Rudra D, et al. Nat Immunol. 2009 Nov;10(11):1170-7. doi: 10.1038/ni.1795. Epub 2009 Sep 20. Nat Immunol. 2009. PMID: 19767756 Free PMC article. - Human T regulatory cells: on the way to cognition.
Kaczorowski M, Jutel M. Kaczorowski M, et al. Arch Immunol Ther Exp (Warsz). 2013 Jun;61(3):229-36. doi: 10.1007/s00005-013-0217-2. Epub 2013 Mar 28. Arch Immunol Ther Exp (Warsz). 2013. PMID: 23536196 Review. - RUNX proteins in transcription factor networks that regulate T-cell lineage choice.
Collins A, Littman DR, Taniuchi I. Collins A, et al. Nat Rev Immunol. 2009 Feb;9(2):106-15. doi: 10.1038/nri2489. Nat Rev Immunol. 2009. PMID: 19165227 Free PMC article. Review.
Cited by
- The immunogenetics of Psoriasis: A comprehensive review.
Harden JL, Krueger JG, Bowcock AM. Harden JL, et al. J Autoimmun. 2015 Nov;64:66-73. doi: 10.1016/j.jaut.2015.07.008. Epub 2015 Jul 26. J Autoimmun. 2015. PMID: 26215033 Free PMC article. Review. - Potential Inhibitory Influence of miRNA 210 on Regulatory T Cells during Epicutaneous Chemical Sensitization.
Long CM, Lukomska E, Marshall NB, Nayak A, Anderson SE. Long CM, et al. Genes (Basel). 2016 Dec 27;8(1):9. doi: 10.3390/genes8010009. Genes (Basel). 2016. PMID: 28035981 Free PMC article. - Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells.
Polansky JK, Schreiber L, Thelemann C, Ludwig L, Krüger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J. Polansky JK, et al. J Mol Med (Berl). 2010 Oct;88(10):1029-40. doi: 10.1007/s00109-010-0642-1. Epub 2010 Jun 24. J Mol Med (Berl). 2010. PMID: 20574810 Free PMC article. - The Roles of RUNX Family Proteins in Development of Immune Cells.
Seo W, Taniuchi I. Seo W, et al. Mol Cells. 2020 Feb 29;43(2):107-113. doi: 10.14348/molcells.2019.0291. Mol Cells. 2020. PMID: 31926543 Free PMC article. Review. - Treg functional stability and its responsiveness to the microenvironment.
Barbi J, Pardoll D, Pan F. Barbi J, et al. Immunol Rev. 2014 May;259(1):115-39. doi: 10.1111/imr.12172. Immunol Rev. 2014. PMID: 24712463 Free PMC article. Review.
References
- Bruno L., Merkenschlager M. 2008. Directing T cell differentiation and function with small molecule inhibitors. Cell Cycle. 7:2296–2298 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials