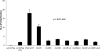Multiple binding modes between HNF4alpha and the LXXLL motifs of PGC-1alpha lead to full activation - PubMed (original) (raw)
Multiple binding modes between HNF4alpha and the LXXLL motifs of PGC-1alpha lead to full activation
Geun Bae Rha et al. J Biol Chem. 2009.
Abstract
Hepatocyte nuclear factor 4alpha (HNF4alpha) is a novel nuclear receptor that participates in a hierarchical network of transcription factors regulating the development and physiology of such vital organs as the liver, pancreas, and kidney. Among the various transcriptional coregulators with which HNF4alpha interacts, peroxisome proliferation-activated receptor gamma (PPARgamma) coactivator 1alpha (PGC-1alpha) represents a novel coactivator whose activation is unusually robust and whose binding mode appears to be distinct from that of canonical coactivators such as NCoA/SRC/p160 family members. To elucidate the potentially unique molecular mechanism of PGC-1alpha recruitment, we have determined the crystal structure of HNF4alpha in complex with a fragment of PGC-1alpha containing all three of its LXXLL motifs. Despite the presence of all three LXXLL motifs available for interactions, only one is bound at the canonical binding site, with no additional contacts observed between the two proteins. However, a close inspection of the electron density map indicates that the bound LXXLL motif is not a selected one but an averaged structure of more than one LXXLL motif. Further biochemical and functional studies show that the individual LXXLL motifs can bind but drive only minimal transactivation. Only when more than one LXXLL motif is involved can significant transcriptional activity be measured, and full activation requires all three LXXLL motifs. These findings led us to propose a model wherein each LXXLL motif has an additive effect, and the multiple binding modes by HNF4alpha toward the LXXLL motifs of PGC-1alpha could account for the apparent robust activation by providing a flexible mechanism for combinatorial recruitment of additional coactivators and mediators.
Figures
FIGURE 1.
Schematic drawings and sequence alignments of the HNF4α and PGC-1α functional regions. A, functional domain structure of HNF4α and the sequence alignment of the LBD of NRs whose crystal structures in complex with the PGC-1α L_XX_LL motifs are known. The residues making contact with the L_XX_LL motif are highlighted in boxes (green and pink), including two charge clamp residues (pink boxes). B, functional domain structure of PGC-1α and its three NR-boxes containing the L_XX_LL motif or its derivatives. Relative positions of the residues comprising the L_XX_LL motif and the flanking regions by the conventional numbering system are shown at the bottom. The characteristic serine and leucine residues of the class III NR-box are also indicated by circles. Regulatory phosphorylation sites are also indicated. These figures are not on the same scale and thus not proportional to the polypeptide chain length of each protein. The abbreviations used are as follows: AF, activation function; AD, activation domain; _NR_s, nuclear receptor boxes, L_XX_LL motifs; ID, inhibitory domain; RS, serine/arginine-rich region; and RRM, RNA recognition motif.
FIGURE 2.
Crystals and the overall structure of HNF4α-PGC-1α complex. A, typical crystals of the complex grown after optimization; B, silver staining of dissolved crystals on SDS-PAGE, which clearly shows the presence of both intact proteins within the crystals. The longest dimension of a typical crystal is around 300 μm. C, Fo − Fc difference electron density map mainly showing the bound L_XX_LL motif and the ligand missing in the initial model. D, ribbon diagram of the final HNF4α-LBD/PGC-1α fragment complex structure. The bound fatty acid is also shown as a _ball_-and-stick model.
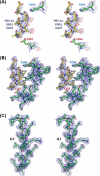
FIGURE 3.
Structural evidence of multiple binding modes by PGC-1α toward HNF4α. A, stereo view of 2_Fo_ − Fc (blue mesh) and Fo − Fc difference maps (red mesh) calculated with the initial model containing a polyalanine model of the bound PGC-1α fragment. The difference map shows the major peaks in the region that only appear in the positions where the invariant leucine residues should be. The charge clamp residues are also shown. B, final 2_Fo_ − Fc map calculated with the refined model containing the leucine residues within the L_XX_LL motifs and the alanine residues for the remaining positions of the PGC-1α fragment. No additional electron densities are visible for PGC-1α side chains. C, final 2_Fo_ − Fc map for one of the core helices of HNF4α as a representative map calculated with a 2.2 Å resolution data set. Side chains can be accurately modeled from this electron density map.
FIGURE 4.
Measurement of transactivation potential exerted by the PGC-1α mutants compared with the wild-type. The 1st 2 lanes refer to empty vectors (1st lane) or transfected with the HNF4α-expressing vector only (2nd lane). w/o, without; w/, with; WT, wild type. The remaining lanes are the results of double transfection of HNF4α- and the respective PGC-1α-expressing vectors. All data have been normalized against the firefly Renilla luciferase activity. Transient transfections were conducted in HeLa cells using HNF4α reporters (_HNF1_α promoters) driving luciferase expression. Values are means ± S.E. (n = 3–4). The mutated NR-boxes are indicated in the name of each mutant. The mutational effects are quite evident, and each mutation causes additional reduction in the overall transcription. Mutations of all three NR-boxes completely ablated the transactivation potential by PGC-1α toward HNF4α.
FIGURE 5.
Set of in vitro binding studies of HNF4α/PGC-1α wild type or mutants. A, GST pulldown with the recombinant proteins of GST-HNF4α-LBD (residues 120–368 containing the hinge region) and the in vitro translated and 35S-labeled PGC-1α wild type (WT) or the mutants (full-length) bearing the mutations within the L_XX_LL motifs. The loading amounts of PGC-1α proteins are indicated in the upper panel, and the retained amounts during the pulldown experiments are shown in the bottom panel. B, ITC analysis of HNF4α against PGC-1α wild-type, mt-NR1+2, and mt-NR1+3 mutants. These three isotherms are shown as representative data, and the complete information on their association constants and the enthalpy and entropy of associations are presented separately in Table 2.
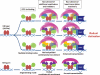
FIGURE 6.
Model scheme of “expanded combinatorial recruitment” for enhanced transactivation. When the coactivator exerts multiple binding modes, it provides a flexible mechanism for combinatorial recruitment and renders an elevated number of opportunities to recruit various additional coactivators and mediators (depicted by various small objects on top of the NR-PGC-1α complex) that can bring the main transcription machinery to the transcription initiation site. Dimeric NRs are shown in red, although PGC-1α in multiple orientations are shown in green. The magenta crescents represent the mediator complex, and the blue objects represent the main transcriptional machinery, including RNA polymerase II (Pol II) and the associated general transcription factors. The remaining small symbols represent various coactivators that can be assembled and some of which have chromatic remodeling activities. This model is focused on the L_XX_LL motif-mediated interactions between PGC-1α and NRs, and the potential involvements of different regions of two proteins for additional interactions are not included.
Similar articles
- Structural and biochemical basis for the binding selectivity of peroxisome proliferator-activated receptor gamma to PGC-1alpha.
Li Y, Kovach A, Suino-Powell K, Martynowski D, Xu HE. Li Y, et al. J Biol Chem. 2008 Jul 4;283(27):19132-9. doi: 10.1074/jbc.M802040200. Epub 2008 May 9. J Biol Chem. 2008. PMID: 18469005 Free PMC article. - Communication between the ERRalpha homodimer interface and the PGC-1alpha binding surface via the helix 8-9 loop.
Greschik H, Althage M, Flaig R, Sato Y, Chavant V, Peluso-Iltis C, Choulier L, Cronet P, Rochel N, Schüle R, Strömstedt PE, Moras D. Greschik H, et al. J Biol Chem. 2008 Jul 18;283(29):20220-30. doi: 10.1074/jbc.M801920200. Epub 2008 Apr 25. J Biol Chem. 2008. PMID: 18441008 - Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha.
Puigserver P. Puigserver P. Int J Obes (Lond). 2005 Mar;29 Suppl 1:S5-9. doi: 10.1038/sj.ijo.0802905. Int J Obes (Lond). 2005. PMID: 15711583 Review. - Nuclear receptor coregulators as a new paradigm for therapeutic targeting.
Hsia EY, Goodson ML, Zou JX, Privalsky ML, Chen HW. Hsia EY, et al. Adv Drug Deliv Rev. 2010 Oct 30;62(13):1227-37. doi: 10.1016/j.addr.2010.09.016. Epub 2010 Oct 7. Adv Drug Deliv Rev. 2010. PMID: 20933027 Free PMC article. Review.
Cited by
- Identification of the Flavonoid Luteolin as a Repressor of the Transcription Factor Hepatocyte Nuclear Factor 4α.
Li J, Inoue J, Choi JM, Nakamura S, Yan Z, Fushinobu S, Kamada H, Kato H, Hashidume T, Shimizu M, Sato R. Li J, et al. J Biol Chem. 2015 Sep 25;290(39):24021-35. doi: 10.1074/jbc.M115.645200. Epub 2015 Aug 13. J Biol Chem. 2015. PMID: 26272613 Free PMC article. - Defining a Canonical Ligand-Binding Pocket in the Orphan Nuclear Receptor Nurr1.
de Vera IMS, Munoz-Tello P, Zheng J, Dharmarajan V, Marciano DP, Matta-Camacho E, Giri PK, Shang J, Hughes TS, Rance M, Griffin PR, Kojetin DJ. de Vera IMS, et al. Structure. 2019 Jan 2;27(1):66-77.e5. doi: 10.1016/j.str.2018.10.002. Epub 2018 Nov 8. Structure. 2019. PMID: 30416039 Free PMC article. - Full-length transcriptomic analysis in murine and human heart reveals diversity of PGC-1α promoters and isoforms regulated distinctly in myocardial ischemia and obesity.
Oehler D, Spychala A, Gödecke A, Lang A, Gerdes N, Ruas J, Kelm M, Szendroedi J, Westenfeld R. Oehler D, et al. BMC Biol. 2022 Jul 30;20(1):169. doi: 10.1186/s12915-022-01360-w. BMC Biol. 2022. PMID: 35907957 Free PMC article. - Synergistic Regulation of Coregulator/Nuclear Receptor Interaction by Ligand and DNA.
de Vera IMS, Zheng J, Novick S, Shang J, Hughes TS, Brust R, Munoz-Tello P, Gardner WJ Jr, Marciano DP, Kong X, Griffin PR, Kojetin DJ. de Vera IMS, et al. Structure. 2017 Oct 3;25(10):1506-1518.e4. doi: 10.1016/j.str.2017.07.019. Epub 2017 Sep 7. Structure. 2017. PMID: 28890360 Free PMC article. - Differential effects, on oncogenic pathway signalling, by derivatives of the HNF4 α inhibitor BI6015.
Kim JH, Eom HJ, Lim G, Park S, Lee J, Nam S, Kim YH, Jeong JH. Kim JH, et al. Br J Cancer. 2019 Mar;120(5):488-498. doi: 10.1038/s41416-018-0374-5. Epub 2019 Feb 22. Br J Cancer. 2019. PMID: 30792535 Free PMC article.
References
- Sladek F. M., Seidel S. D. (2001) in Nuclear Receptors in Genetic Disease (Burris T. P., McCabe E. R. B., eds) Academic Press, New York
- Parviz F., Matullo C., Garrison W. D., Savatski L., Adamson J. W., Ning G., Kaestner K. H., Rossi J. M., Zaret K. S., Duncan S. A. (2003) Nat. Genet. 34, 292–296 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous