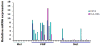Distinct MHC gene expression patterns during progression of melanoma - PubMed (original) (raw)
Distinct MHC gene expression patterns during progression of melanoma
Yan Degenhardt et al. Genes Chromosomes Cancer. 2010 Feb.
Abstract
Abnormal expression of major histocompatibility complex (MHC) molecules in melanoma has been reported previously. However, the MHC molecule expression patterns in different growth phases of melanoma and the underlying mechanisms are not well understood. Here, we demonstrate that in vertical growth phase (VGP) melanomas, MHC genes are subject to increased rates of DNA copy number gains, accompanied by increased expression, in comparison to normal melanocytes. In contrast, MHC expression in metastatic melanomas drastically decreased compared to VGP melanomas, despite still prevalent DNA copy number gains. Subsequent investigations found that the master transactivator of MHC genes, CIITA, was also significantly downregulated in metastatic melanomas when compared to VGP melanomas. This could be one of the mechanisms accounting for the discrepancy between DNA copy number and expression level in metastatic melanomas, a potentially separate mechanism of gene regulation. These results infer a dynamic role of MHC function in melanoma progression. We propose potential mechanisms for the overexpression of MHC molecules in earlier stages of melanoma as well as for its downregulation in metastatic melanomas.
Figures
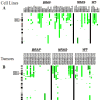
Figure 1
Copy number aberrations of MHC molecules in melanoma cells. Copy number gains of _MHC_-I and _MHC_-II (6p21.32) are common in melanomas. Vertical green bars represent the extent of DNA copy number gains along chromosome 6 in a panel of 52 cancer cell lines (A) and 43 tumor tissues (B). With respect to the copy number changes, the cell lines are good models for the tumor tissues, where the copy number gain frequency is 32.7% and 25.6% respectively. Only 3 samples (1 cell line, 2 tumors) harbored DNA gains of > ~5 copies (dark green).
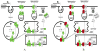
Figure 2
Aberrant expression of MHC molecules in VGP melanoma cell lines and metastatic melanoma cell lines compared to melanocytes. (A) Aberrant expression of MHC molecules in the antigen presentation pathway in VGP melanoma cells compared to melanocytes viewed with the Ingenuity pathway analysis software. Significantly differentially expressed genes (_P_-value < 0.05) with fold change over 1.5 between VGP cells and melanocytes were loaded into and viewed with the Ingenuity pathway analysis software. Green represents genes with increased expression and red represents genes with decreased expression compared to melanocytes. (B) Aberrant expression of MHC molecules in the antigen presentation pathway in metastatic melanoma cells compared to melanocytes viewed with the Ingenuity pathway analysis software. Significantly differentially expressed genes (_P_-value < 0.05) with fold change over 1.5 between metastatic cells and melanocytes were loaded into and viewed with the Ingenuity pathway analysis software. Green represents genes with increased expression and red represents genes with decreased expression compared to melanocytes.
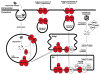
Figure 3
Significant decrease of gene expression in the antigen presentation pathway in metastatic melanoma cell lines in comparison to VGP melanoma cell lines. Significantly differentially expressed genes (_P_-value < 0.05) between metastatic and VGP melanoma cell lines, also with fold change over 1.5, were loaded into and viewed with the Ingenuity pathway analysis software. Red represents decreased expression of components of this pathway in metastatic cells compared to VGP cells.
Figure 4
Taqman gene expression assay and Western blot show that expression levels for HLA-DRA and CD74 are non-detectable in melanocytes and very low in metastatic cell lines, but are strongly elevated in VGP melanoma cell lines. HLA-DRA (A) and CD74 (B) mRNA levels were measured by rt-taqman in a group of melanocyte cell lines, VGP lines and metastatic melanoma cell lines, and normalized to β-actin house-keeping gene mRNA level. The result presented here is the average of two independent data points from a single experiment. The same experiment was done twice with similar results. The names of the cell lines in the Figures are listed in Table 4 in the same order. The protein extracts were made from a subset of these cell lines and probed for HLA-DRA and CD74 protein levels by western. GAPDH was used as loading control.
Figure 5
Taqman gene expression assay shows that in concordance with HLA-DRA expression, CIITA is highly expressed in VGP cell lines, but is non-detectable in melanocytes and expressed at very low level in metastatic cell lines. HLA-DRA and CIITA mRNA levels were measured by rt-taqman, and relative expressions to β-actin were plotted side-by-side for the same cell lines. The result presented here is the average of two independent data points from a single experiment. The same experiment was done twice with similar results. The names of the cell lines in the Figure are listed in Table 4 in the same order.
Similar articles
- Class II transactivator (CIITA) isoform expression and activity in melanoma.
Baton F, Deruyffelaere C, Chapin M, Prod'homme T, Charron D, Al-Daccak R, Alcaide-Loridan C. Baton F, et al. Melanoma Res. 2004 Dec;14(6):453-61. doi: 10.1097/00008390-200412000-00004. Melanoma Res. 2004. PMID: 15577315 - Epigenetic silencing of the CIITA gene and posttranscriptional regulation of class II MHC genes in ocular melanoma cells.
Radosevich M, Song Z, Gorga JC, Ksander B, Ono SJ. Radosevich M, et al. Invest Ophthalmol Vis Sci. 2004 Sep;45(9):3185-95. doi: 10.1167/iovs.04-0111. Invest Ophthalmol Vis Sci. 2004. PMID: 15326139 - cDNA-array profiling of melanomas and paired melanocyte cultures.
Mischiati C, Natali PG, Sereni A, Sibilio L, Giorda E, Cappellacci S, Nicotra MR, Mariani G, Di Filippo F, Catricalà C, Gambari R, Grammatico P, Giacomini P. Mischiati C, et al. J Cell Physiol. 2006 Jun;207(3):697-705. doi: 10.1002/jcp.20610. J Cell Physiol. 2006. PMID: 16523488 - Gene regulation in melanoma progression by the AP-2 transcription factor.
Bar-Eli M. Bar-Eli M. Pigment Cell Res. 2001 Apr;14(2):78-85. doi: 10.1034/j.1600-0749.2001.140202.x. Pigment Cell Res. 2001. PMID: 11310795 Review. - Transcriptional regulation of metastasis-related genes in human melanoma.
Nyormoi O, Bar-Eli M. Nyormoi O, et al. Clin Exp Metastasis. 2003;20(3):251-63. doi: 10.1023/a:1022991302172. Clin Exp Metastasis. 2003. PMID: 12741683 Review.
Cited by
- Genomic landscape of the immunogenicity regulation in skin melanomas with diverse tumor mutation burden.
Georgoulias G, Zaravinos A. Georgoulias G, et al. Front Immunol. 2022 Oct 28;13:1006665. doi: 10.3389/fimmu.2022.1006665. eCollection 2022. Front Immunol. 2022. PMID: 36389735 Free PMC article. - HLA-DRB1: A new potential prognostic factor and therapeutic target of cutaneous melanoma and an indicator of tumor microenvironment remodeling.
Deng H, Chen Y, Wang J, An R. Deng H, et al. PLoS One. 2022 Sep 21;17(9):e0274897. doi: 10.1371/journal.pone.0274897. eCollection 2022. PLoS One. 2022. PMID: 36129956 Free PMC article. - The Transcriptional Landscape of BRAF Wild Type Metastatic Melanoma: A Pilot Study.
Lastraioli E, Ruffinatti FA, Bagni G, Visentin L, di Costanzo F, Munaron L, Arcangeli A. Lastraioli E, et al. Int J Mol Sci. 2022 Jun 21;23(13):6898. doi: 10.3390/ijms23136898. Int J Mol Sci. 2022. PMID: 35805902 Free PMC article. - The Role of Extracellular Matrix Remodeling in Skin Tumor Progression and Therapeutic Resistance.
Fromme JE, Zigrino P. Fromme JE, et al. Front Mol Biosci. 2022 Apr 26;9:864302. doi: 10.3389/fmolb.2022.864302. eCollection 2022. Front Mol Biosci. 2022. PMID: 35558554 Free PMC article. Review. - Distinct Molecular Mechanisms of Altered HLA Class II Expression in Malignant Melanoma.
Meyer S, Handke D, Mueller A, Biehl K, Kreuz M, Bukur J, Koehl U, Lazaridou MF, Berneburg M, Steven A, Massa C, Seliger B. Meyer S, et al. Cancers (Basel). 2021 Aug 3;13(15):3907. doi: 10.3390/cancers13153907. Cancers (Basel). 2021. PMID: 34359808 Free PMC article.
References
- Altomonte M, Fonsatti E, Visintin A, Maio M. Targeted therapy of solid malignancies via HLA class II antigens: a new biotherapeutic approach? Oncogene. 2003;22:6564–6569. - PubMed
- Anichini A, Mortarini R, Nonaka D, Molla A, Vegetti C, Montaldi E, Wang X, Ferrone S. Association of antigen-processing machinery and HLA antigen phenotype of melanoma cells with survival in American Joint Committee on Cancer stage III and IV melanoma patients. Cancer Res. 2006;66:6405–6411. - PubMed
- Aoudjit F, Guo W, Gagnon-Houde JV, Castaigne JG, Alcaide-Loridan C, Charron D, Al-Daccak R. HLA-DR signaling inhibits Fas-mediated apoptosis in A375 melanoma cells. Exp Cell Res. 2004;299:79–90. - PubMed
- Balazs M, Adam Z, Treszl A, Begany A, Hunyadi J, Adany R. Chromosomal imbalances in primary and metastatic melanomas revealed by comparative genomic hybridization. Cytometry. 2001;46:222–232. - PubMed
- Byers HR, Bhawan J. Pathologic parameters in the diagnosis and prognosis of primary cutaneous melanoma. Hematol Oncol Clin North Am. 1998;12:717–735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials