Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production - PubMed (original) (raw)
. 2009 Dec;10(12):1283-91.
doi: 10.1038/ni.1820. Epub 2009 Nov 8.
Teresa Lambe, Andy L Johnson, Bebhinn Treanor, Edyta Kucharska, Heather Domaschenz, Belinda Whittle, Lina E Tze, Anselm Enders, Tanya L Crockford, Tiphaine Bouriez-Jones, Duncan Alston, Jason G Cyster, Michael J Lenardo, Fabienne Mackay, Elissa K Deenick, Stuart G Tangye, Tyani D Chan, Tahra Camidge, Robert Brink, Carola G Vinuesa, Facundo D Batista, Richard J Cornall, Christopher C Goodnow
Affiliations
- PMID: 19898472
- PMCID: PMC3437189
- DOI: 10.1038/ni.1820
Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production
Katrina L Randall et al. Nat Immunol. 2009 Dec.
Erratum in
- Nat Immunol. 2010 Jul;11(7):644. Johnson, Andy [corrected to Johnson, Andy L]
Abstract
To identify genes and mechanisms involved in humoral immunity, we did a mouse genetic screen for mutations that do not affect the first wave of antibody to immunization but disrupt response maturation and persistence. The first two mutants identified had loss-of-function mutations in the gene encoding a previously obscure member of a family of Rho-Rac GTP-exchange factors, DOCK8. DOCK8-mutant B cells were unable to form marginal zone B cells or to persist in germinal centers and undergo affinity maturation. Dock8 mutations disrupted accumulation of the integrin ligand ICAM-1 in the B cell immunological synapse but did not alter other aspects of B cell antigen receptor signaling. Humoral immunodeficiency due to Dock8 mutation provides evidence that organization of the immunological synapse is critical for signaling the survival of B cell subsets required for long-lasting immunity.
Figures
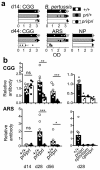
Figure 1. Discovery of two independent, non-complementing ENU mutant mouse strains with humoral immunodeficiency characterized by failure to sustain a primary immune response and failure of maturation of the antibody response
(a) Representative screening assay results. IgG1 antibody to CGG and ARS, IgM antibody to NP, and IgG2c antibody to B. pertussis measured by ELISA optical density (OD) in serum collected 14 days post-primary immunization (d14) and 6 days after booster immunization (d44). To detect low responders among large numbers of mice, serum was tested at a fixed dilution yielding sub-saturating antibody titers in wild-type animals (CGG 1/2000, 1/20,000; BP 1/50; NP 1/50; ABA 1/2000). (b) Antibody titers to CGG and ARS followed longitudinally 14, 28 and 56 days after primary immunization, determined by titrating each serum sample against a standard reference serum whose titer was set as 1. Statistical comparison by Student’s _t_-test: ns, not significant; * P < 0.05; ** P < 0.005; *** P < 0.0005). In a,b, columns are arithmetic means, bars standard error of the mean (s.e.m), and dots individual mice. Data are representative of 3 independent experiments.
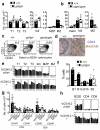
Figure 2. Analysis of splenic and peritoneal lymphocyte subsets
(a, b) Number of immature transitional 1-3 B cells (T1-T3, Trans), mature follicular B cells (Foll), marginal zone B cell precursors (MZP) and marginal zone B cells (MZ) in spleen of individual mice of the indicated genotypes. MZ B cell numbers were statistically significant (unpaired, two-tailed t test, (a) P = 0.006, (b) P = 0.038), while other B subsets were not significantly different. Data representative of 2 independent experiments. (c) Representative plots (from >5 independent experiments) showing % of B cells in the MZ B cell subset. (d) Immunohistochemistry of splenic MZ B cell marker CD1d and marginal sinus marker MAdCAM. (e) Percentage of mutant or wild-type CD45.2+ cells reconstituting the indicated B cell subsets in the spleen of individual mixed chimera mice. Differences in MZ contribution are representative of two separate cohorts of pri/pri mixed chimeras and two cohorts of cpm/cpm mixed chimeras, while no consistent difference in contribution to follicular or transitional cells was observed. (f) Numbers of B1, B1a, B1b and B2 subsets in the peritoneum of mice of the indicated genotypes. Representative of two independent experiments. (g) Numbers of splenic T cells in individual mice of the indicated genotypes. Naïve T cell numbers (CD44lo) were statistically significantly different (ANOVA) for both CD4+ (P = 0.0003) and CD8+ T cells (P = 0.0001). Differences in CD8CD44hi cells were statistically significantly different (P = 0.048), but not in CD4CD44hi. (h) Percentage of mutant or wild-type CD45.2+ cells reconstituting the indicated splenic cell subsets of individual mixed chimeras. Representative of three independent experiments.
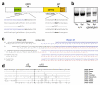
Figure 3. Independent Dock8 mutations in pri and cpm mouse strains
(a) Conserved DOCK8 domains and mutations. (b) Dock8 exon 18-21 cDNA amplification products from thymus (Thy) and spleen (Spl) of wild-type and cpm mice. Data are representative of 4 independent experiments. (c) Sequence of cDNA clones from cpm/cpm mice in (b), aligned to wild-type Dock8 cDNA sequence. Number of clones of each type shown on left. //, segment of normal exon 20 sequence omitted. (d) Sequence alignment of DOCK8 orthologues and paralogues in the region surrounding the pri mutation. Mutated serine in bold; dashes indicate identity to mouse DOCK8; secondary structure elements based on the structure of DOCK9 ; triangles show Cdc42 contact residues in DOCK9 .
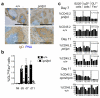
Figure 4. DOCK8 mutation causes an intrinsic defect in germinal center B cells
(a) Immunohistochemistry of splenic PNA+ germinal centers and IgD+ follicular mantle 7 days after SRBC-immunization (scale bars: upper 500μm; lower 200μm). (b) Enumeration of GC B cells as percentage of B220+ in untreated mice (nil) or 5, 7 and 11 days after SRBC-immunization. Columns are arithmetic means, bars standard error of the mean (s.e.m), and dots individual mice (c) Contribution to different B cell subsets in 50:50 mixed bone marrow chimeras 7 or 11 days after SRBC immunization. Columns show percentage of CD45.2+ cells of the indicated genotypes in individual, numbered chimeric mice. Each bar represents an individual mouse. Numbers indicate total number of GC cells ×106 per spleen (mean+/−SD) in each group of mice. Data are from 3 independent experiments.
Figure 5. DOCK8 mutant B cells undergo normal T-cell dependent activation, switching and initial differentiation into GC cells in vivo
HEL-specific pri/pri or wild-type (+/+) B cells from SWHEL transgenic mice, allotypically tagged by CD45.1, were adoptively transferred into wild-type mice that were immunized with HEL2X-SRBC and analyzed on the indicated days after transfer and immunization. (a) Representative flow cytometric analyses of spleen gated on HEL+ CD45.1+ B220+ SWHEL B cells, showing percentage of GL7+ Fas+ cells (left) and IgG1+ switched and unswitched cells (right). Control mice given SWHEL B cells but immunized with unconjugated SRBC lacking HEL (SRBC) are marked (*). (b) Enumeration of number of HEL+ CD45.1+ B220+ SWHEL B cells on the indicated days after immunization, and the percentage of SWHEL B cells that were GL7+ Fas+ or switched to IgG1 in individual animals. Statistical analysis (wild-type vs. pri/pri) by unpaired _t_-test with Welch’s correction. Unfilled circles represent wild-type, filled circles pri/pri and (*) control srbc mice. Early time-course experiment performed once.
Figure 6. Intrinsic failure of DOCK8 mutant germinal center B cells to persist or undergo affinity maturation
HEL-specific pri/pri or wild-type (+/+) B cells from SWHEL transgenic mice, allotypically tagged by CD45.1, were injected into wild-type B6 mice and the recipients immunized with HEL2X-SRBC. (a) Confocal immunofluorescence of spleen cryosections from recipient mice 5 or 10 days after immunization, stained for HEL, IgD and CD3. GCs (arrows) and extrafollicular plasma cell foci (PC), and T cell zones (T) are marked. (b) Representative flow cytometry plots and enumeration of CD45.1+ HEL-binding B cells and GC cells derived from the HEL+ CD45.1+ SWHEL cells, of the indicated genotypes, 5-9 days after adoptive transfer. Control mice given SWHEL B cells but immunized with unconjugated SRBC without HEL (srbc) are marked (*). Panels also show total number of GC B cells per spleen in individual mice and the percentage of GC B cells that were SWHEL B cells (day 9, +/+ vs. pri/pri, P = 0.008) and the percentage and absolute numbers of HEL+ CD45.1+ SWHEL FAShi HELlo GC cells and IgG1-switched cells. Unfilled circles represent wild-type, filled circles pri/pri and (*) control srbc mice. (c) Structure of HEL complexed with the SWHEL antibody HyHEL10 showing the position of the two HEL residues mutated in HEL2X and four of the D101 contact residues in the heavy chain CDR2. (d) Results of sequencing of heavy chain genes from individual flow-sorted SWHEL GC B cells (both +/+ or pri/pri). Graphs show the percentage of cells containing the indicated number of mutations in the CDR2; the total number of cells sequenced is indicated in the center. The replacement to silent ratio (R/S) and percentage of cells with a Y58F replacement mutation are also shown. (e) Enumeration of high affinity variants on day 9 post-immunization by direct assay for binding to nanomolar concentrations of D101R mutant HEL3X (independent experiment from b, d). Flow cytometry plots, gated on B220+ CD45.1+ SWHEL B cells, are from concatenated data collected on 11-22 million spleen cells combined from four individual recipients of either +/+ or pri/pri SWHEL B cells or from two recipients immunized with uncoupled SRBC (*). The mean % of spleen cells that are CD45.1+ SWHEL B cells is shown in blue and the mean % of spleen cells switched to IgG1 is shown in red. Staining with nanomolar HEL3X reveals mutated B cells with higher affinity BCRs (oval gate), and the mean % spleen cells in this gate shown in black. Panels on right show data for individual animals in each group. Statistical analysis by Student’s t-test with Welch’s correction indicated a significant difference between pri/pri and +/+ SWHEL cells in percentage of high affinity cells among either CD45.1+ B220+ cells (P = 0.038) or among CD45.1+ B220+ IgG1+ cells (P = 0.031) that remained on day 9. Data are representative of four independent experiments, except (d) and (e) which were each performed once but independently of each other.
Figure 7. DOCK8 mutations disrupt the formation of the B cell immunological synapse but not other aspects of signaling through the B cell antigen receptor
(a) HEL-specific MD4 wild-type or mutant B cells were settled onto lipid bilayers containing mono-biotinylated HEL (green) and Alexa-532-conjugated ICAM-1 (red) and imaged after 10 min of interaction. Differential interference contrast (DIC), confocal fluorescence for HEL and ICAM-1, and interference reflection microscopy (IRM) images of representative cells are shown. Right, quantification of the area of B cell contact with the bilayer, the relative amounts of antigen accumulated at the contact site, and the percentages of B cells forming a peripheral ring of ICAM-1 (pSMAC). Results representative of 3 independent experiments. Columns are means and error bars are s.e.m. (b) Intracellular calcium in mixtures of pri/pri CD45.2 (red trace) and wild-type CD45.1 spleen cells stimulated with anti-IgM (arrow). (c) Expression of CD25 and CD86 on lymph node B cells 18 h after stimulation with 1 or 10 μg/ml anti-IgM. Unstimulated, grey histogram; wild-type, black line; pri/pri, red filled. Representative plots from 4 independent mice. (d) Division of LN B cells measured by dilution of CFSE in B220+ 7AAD− cells after 4 d culture with the indicated stimuli. Mutant pri/pri or cpm/cpm CD45.2 cells (red histograms) were CFSE labeled and cultured with wild-type CD45.1 cells (black histograms) in the same wells to control for experimental variation. Representative of 6 out of 8 independent samples tested: in 2 samples there was a subtle decrease in CFSE dilution by pri/pri cells (Supplementary Fig. 9).
Comment in
- B cell memory: how to start and when to end.
Pelletier N, McHeyzer-Williams MG. Pelletier N, et al. Nat Immunol. 2009 Dec;10(12):1233-5. doi: 10.1038/ni1209-1233. Nat Immunol. 2009. PMID: 19915622 No abstract available.
Similar articles
- B cells require DOCK8 to elicit and integrate T cell help when antigen is limiting.
Deobagkar-Lele M, Crawford G, Crockford TL, Back J, Hodgson R, Bhandari A, Bull KR, Cornall RJ. Deobagkar-Lele M, et al. Sci Immunol. 2024 Aug 9;9(98):eadd4874. doi: 10.1126/sciimmunol.add4874. Epub 2024 Aug 9. Sci Immunol. 2024. PMID: 39121196 Free PMC article. - DOCK8 is essential for LFA-1-dependent positioning of T follicular helper cells in germinal centers.
Janssen E, Tohme M, Butts J, Giguere S, Sage PT, Velázquez FE, Kam C, Milin E, Das M, Sobh A, Al-Tamemi S, Luscinskas FW, Batista F, Geha RS. Janssen E, et al. JCI Insight. 2020 Aug 6;5(15):e134508. doi: 10.1172/jci.insight.134508. JCI Insight. 2020. PMID: 32573493 Free PMC article. - The essential role of DOCK8 in humoral immunity.
Randall KL, Lambe T, Goodnow CC, Cornall RJ. Randall KL, et al. Dis Markers. 2010;29(3-4):141-50. doi: 10.3233/DMA-2010-0739. Dis Markers. 2010. PMID: 21178273 Free PMC article. Review. - DOCK8 gene mutation alters cell subsets, BCR signaling, and cell metabolism in B cells.
Gu H, Xie M, Zhao S, Luo X, Huang Y, Yang L, Guan F, Lei J, Liu C. Gu H, et al. Cell Death Dis. 2024 Dec 1;15(11):871. doi: 10.1038/s41419-024-07180-w. Cell Death Dis. 2024. PMID: 39616183 Free PMC article. - T cell interactions with B cells during germinal center formation, a three-step model.
Biram A, Davidzohn N, Shulman Z. Biram A, et al. Immunol Rev. 2019 Mar;288(1):37-48. doi: 10.1111/imr.12737. Immunol Rev. 2019. PMID: 30874355 Review.
Cited by
- Sequential class switching is required for the generation of high affinity IgE antibodies.
Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA, Lafaille JJ. Xiong H, et al. J Exp Med. 2012 Feb 13;209(2):353-64. doi: 10.1084/jem.20111941. Epub 2012 Jan 16. J Exp Med. 2012. PMID: 22249450 Free PMC article. - Dedicator of cytokinesis 8 interacts with talin and Wiskott-Aldrich syndrome protein to regulate NK cell cytotoxicity.
Ham H, Guerrier S, Kim J, Schoon RA, Anderson EL, Hamann MJ, Lou Z, Billadeau DD. Ham H, et al. J Immunol. 2013 Apr 1;190(7):3661-9. doi: 10.4049/jimmunol.1202792. Epub 2013 Mar 1. J Immunol. 2013. PMID: 23455509 Free PMC article. - Immunity and Genetics at the Revolving Doors of Diagnostics in Primary Immunodeficiencies.
Rispoli F, Valencic E, Girardelli M, Pin A, Tesser A, Piscianz E, Boz V, Faletra F, Severini GM, Taddio A, Tommasini A. Rispoli F, et al. Diagnostics (Basel). 2021 Mar 16;11(3):532. doi: 10.3390/diagnostics11030532. Diagnostics (Basel). 2021. PMID: 33809703 Free PMC article. - The Hyper-IgE Syndromes: Lessons in Nature, From Bench to Bedside.
Rael EL, Marshall RT, McClain JJ. Rael EL, et al. World Allergy Organ J. 2012 Jul;5(7):79-87. doi: 10.1097/WOX.0b013e31825a73b2. World Allergy Organ J. 2012. PMID: 23283142 Free PMC article. - Non-additive QTL mapping of lactation traits in 124,000 cattle reveals novel recessive loci.
Reynolds EGM, Lopdell T, Wang Y, Tiplady KM, Harland CS, Johnson TJJ, Neeley C, Carnie K, Sherlock RG, Couldrey C, Davis SR, Harris BL, Spelman RJ, Garrick DJ, Littlejohn MD. Reynolds EGM, et al. Genet Sel Evol. 2022 Jan 24;54(1):5. doi: 10.1186/s12711-021-00694-3. Genet Sel Evol. 2022. PMID: 35073835 Free PMC article.
References
- Conley ME, et al. Primary B cell immunodeficiencies: comparisons and contrasts. Annu Rev Immunol. 2009;27:199–227. - PubMed
- Park MA, Li JT, Hagan JB, Maddox DE, Abraham RS. Common variable immunodeficiency: a new look at an old disease. Lancet. 2008;372:489–502. - PubMed
- Schaffer AA, Salzer U, Hammarstrom L, Grimbacher B. Deconstructing common variable immunodeficiency by genetic analysis. Curr Opin Genet Dev. 2007;17:201–212. - PubMed
- van Zelm MC, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. New England Journal of Medicine. 2006;354:1901–1912. - PubMed
- Tuveson DA, Carter RH, Soltoff SP, Fearon DT. CD19 of B cells as a surrogate kinase insert region to bind phosphatidylinositol 3-kinase. Science. 1993;260:986–989. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0901117/MRC_/Medical Research Council/United Kingdom
- R01 AI052127/AI/NIAID NIH HHS/United States
- 062920/WT_/Wellcome Trust/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
- 082030/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous