PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification - PubMed (original) (raw)
. 2010 Jan;38(Database issue):D792-9.
doi: 10.1093/nar/gkp1005. Epub 2009 Nov 11.
Affiliations
- PMID: 19906719
- PMCID: PMC2808898
- DOI: 10.1093/nar/gkp1005
PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification
Athanasia Spandidos et al. Nucleic Acids Res. 2010 Jan.
Abstract
PrimerBank (http://pga.mgh.harvard.edu/primerbank/) is a public resource for the retrieval of human and mouse primer pairs for gene expression analysis by PCR and Quantitative PCR (QPCR). A total of 306,800 primers covering most known human and mouse genes can be accessed from the PrimerBank database, together with information on these primers such as T(m), location on the transcript and amplicon size. For each gene, at least one primer pair has been designed and in many cases alternative primer pairs exist. Primers have been designed to work under the same PCR conditions, thus facilitating high-throughput QPCR. There are several ways to search for primers for the gene(s) of interest, such as by: GenBank accession number, NCBI protein accession number, NCBI gene ID, PrimerBank ID, NCBI gene symbol or gene description (keyword). In all, 26,855 primer pairs covering most known mouse genes have been experimentally validated by QPCR, agarose gel analysis, sequencing and BLAST, and all validation data can be freely accessed from the PrimerBank web site.
Figures
Figure 1.
PrimerBank search results for beta-actin primers. The primer search function can be found on the PrimerBank homepage. The database was searched for mouse beta-actin primer pairs by PrimerBank ID (6671509a1) and the search results obtained are shown here. The primer sequences, lengths, _T_ms, location of primers on the amplicon and expected amplicon size can be seen.
Figure 2.
cDNA and amplicon sequence for beta-actin primers. The full cDNA and amplicon sequences as well as the highlighted location of primers on the amplicon are seen here and can be viewed from PrimerBank by clicking on the cDNA and amplicon sequence link found on the primer information web page (seen in Figure 1).
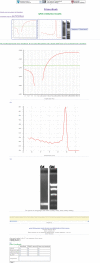
Figure 3.
Experimental validation data for beta-actin primers. All experimental validation data are seen here and can be viewed from PrimerBank by clicking on the validation results links found on the primer information web page (seen in Figure 1) and on the cDNA and amplicon sequence web page (seen in Figure 2). The QPCR amplification plot, the dissociation curve and agarose gel data can be seen followed by the sequence obtained, which users can scroll through. A summary of the BLAST results obtained can be seen below the sequencing result. A BLAST tool can be seen below the BLAST results, which can be used to directly BLAST the sequence obtained against the NCBI database. Also, a feedback table can be seen, where users can provide information about their experimental data using the primers.
Similar articles
- PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update.
Wang X, Spandidos A, Wang H, Seed B. Wang X, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D1144-9. doi: 10.1093/nar/gkr1013. Epub 2011 Nov 15. Nucleic Acids Res. 2012. PMID: 22086960 Free PMC article. - A PCR primer bank for quantitative gene expression analysis.
Wang X, Seed B. Wang X, et al. Nucleic Acids Res. 2003 Dec 15;31(24):e154. doi: 10.1093/nar/gng154. Nucleic Acids Res. 2003. PMID: 14654707 Free PMC article. - methBLAST and methPrimerDB: web-tools for PCR based methylation analysis.
Pattyn F, Hoebeeck J, Robbrecht P, Michels E, De Paepe A, Bottu G, Coornaert D, Herzog R, Speleman F, Vandesompele J. Pattyn F, et al. BMC Bioinformatics. 2006 Nov 9;7:496. doi: 10.1186/1471-2105-7-496. BMC Bioinformatics. 2006. PMID: 17094804 Free PMC article. - FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents.
Hu Y, Sopko R, Foos M, Kelley C, Flockhart I, Ammeux N, Wang X, Perkins L, Perrimon N, Mohr SE. Hu Y, et al. G3 (Bethesda). 2013 Sep 4;3(9):1607-16. doi: 10.1534/g3.113.007021. G3 (Bethesda). 2013. PMID: 23893746 Free PMC article. - Graphical design of primers with PerlPrimer.
Marshall O. Marshall O. Methods Mol Biol. 2007;402:403-14. doi: 10.1007/978-1-59745-528-2_21. Methods Mol Biol. 2007. PMID: 17951808 Review.
Cited by
- N6-methyladenosine of Socs1 modulates macrophage inflammatory response in different stiffness environments.
Hu Z, Li Y, Yuan W, Jin L, Leung WK, Zhang C, Yang Y. Hu Z, et al. Int J Biol Sci. 2022 Sep 11;18(15):5753-5769. doi: 10.7150/ijbs.74196. eCollection 2022. Int J Biol Sci. 2022. PMID: 36263168 Free PMC article. - Epithelial-mesenchymal transition induction is associated with augmented glucose uptake and lactate production in pancreatic ductal adenocarcinoma.
Liu M, Quek LE, Sultani G, Turner N. Liu M, et al. Cancer Metab. 2016 Oct 17;4:19. doi: 10.1186/s40170-016-0160-x. eCollection 2016. Cancer Metab. 2016. PMID: 27777765 Free PMC article. - Gene expression analysis of the pleiotropic effects of TGF-β1 in an in vitro model of flexor tendon healing.
Farhat YM, Al-Maliki AA, Chen T, Juneja SC, Schwarz EM, O'Keefe RJ, Awad HA. Farhat YM, et al. PLoS One. 2012;7(12):e51411. doi: 10.1371/journal.pone.0051411. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251524 Free PMC article. - Stimulation of regulatory T cells with Lactococcus lactis expressing enterotoxigenic E. coli colonization factor antigen 1 retains salivary flow in a genetic model of Sjögren's syndrome.
Akgul A, Maddaloni M, Jun SM, Nelson AS, Odreman VA, Hoffman C, Bhagyaraj E, Voigt A, Abbott JR, Nguyen CQ, Pascual DW. Akgul A, et al. Arthritis Res Ther. 2021 Apr 6;23(1):99. doi: 10.1186/s13075-021-02475-1. Arthritis Res Ther. 2021. PMID: 33823920 Free PMC article. - Psychostimulants Modafinil, Atomoxetine and Guanfacine Impair Bone Cell Differentiation and MSC Migration.
Wagener N, Lehmann W, Weiser L, Jäckle K, Di Fazio P, Schilling AF, Böker KO. Wagener N, et al. Int J Mol Sci. 2022 Sep 6;23(18):10257. doi: 10.3390/ijms231810257. Int J Mol Sci. 2022. PMID: 36142172 Free PMC article.
References
- Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. - PubMed
- Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR – a perspective. J. Mol. Endocrinol. 2005;34:597–601. - PubMed
- Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. - PubMed
- VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials