Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer - PubMed (original) (raw)
Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer
Chunru Lin et al. Cell. 2009.
Abstract
Chromosomal translocations are a hallmark of leukemia/lymphoma and also appear in solid tumors, but the underlying mechanism remains elusive. By establishing a cellular model that mimics the relative frequency of authentic translocation events without proliferation selection, we report mechanisms of nuclear receptor-dependent tumor translocations. Intronic binding of liganded androgen receptor (AR) first juxtaposes translocation loci by triggering intra- and interchromosomal interactions. AR then promotes site-specific DNA double-stranded breaks (DSBs) at translocation loci by recruiting two types of enzymatic activities induced by genotoxic stress and liganded AR, including activation-induced cytidine deaminase and the LINE-1 repeat-encoded ORF2 endonuclease. These enzymes synergistically generate site-selective DSBs at juxtaposed translocation loci that are ligated by nonhomologous end joining pathway for specific translocations. Our data suggest that the confluence of two parallel pathways initiated by liganded nuclear receptor and genotoxic stress underlies nonrandom tumor translocations, which may function in many types of tumors and pathological processes.
Figures
Figure 1. Liganded-AR and Genotoxic Stress Synergistically Induce Chromosomal Translocations in Prostate Cancer Cells
(A and B) Identification and characterization of induced TMPRSS2:ERGb and TMPRSS2:ETV1b translocations in LNCaP cells. Top: schematic structures for the TMPRSS2, ERG, and ETV1 mRNA indicating exon positions. Middle: RT-PCR amplification of TMPRSS2:ERGb (A) or TMPRSS2:ETV1b (B) fusion transcripts from 48 individual cell samples. Bottom: confirmation of position and fusion sites by automated DNA sequencing. (C and D) Statistical analysis of DHT and IR induced TMPRSS2:ERG (C) and TMPRSS2:ETV1 (D) translocations (_n_=3, ±SEM). (E and F) Identification and characterization of induced TMPRSS2:ERG (E) and TMPRSS2:ETV1 (F) translocation isoforms in LNCaP cells. Bottom: summary of distinct fusion types. (G and H) Involvement of DNA repair machinery in induced TMPRSS2:ERGb and TMPRSS2:ETV1b translocations by QPCR with indicated siRNAs (_n_=3, ±SEM).
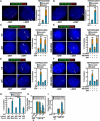
Figure 2. AR-Induced and Motor-Dependent Chromosomal Interactions of TMPRSS2 and ERG or ETV1 Loci
(A and B) Interphase FISH analysis on PrEC cells with TMPRSS2 (green), ERG (red) (A), or ETV1 (red) (B) probes. (C and D) Actin polymerization-dependent interchromosomal interactions. (E and F) Nuclear myosin-dependent interchromosomal interactions. (G) ATPase activity of NMI is required for DHT induced interchromosomal interactions. (H and I) Requirement of nuclear myosin/actin motor system for induced TMPRSS2:ERGb and TMPRSS2:ETV1b translocations in LNCaP cells pretreated with Latrunculin (LtA) or Jasplakinolide (Jpk) (H) or transfected with NMI siRNA (I) (_n_=3, ±SEM).
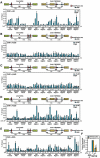
Figure 3. Identification of Breakpoints for TMPRSS2:ERG and TMPRSS2:ETV1 Translocations
(A) The tracks display the human genome coordinates (hg18 assembly), red band: predicted potential DSBs based on ChIP-seq using anti-BrdU antibodies (see Experimental Procedures). Boxes: exons; lines: introns; Region I to IV: DNA break points within corresponding loci. (B) The fold change of the tag density in the double-strand break regions I, II, III and IV after DHT treatment. (C and D) Conventional ChIP analysis with anti-BrdU antibodies on ERG and TMPRSS2 intronic break regions identified by ChIP-seq. (E-G) Identification and characterization of induced TMPRSS2:ERG and TMPRSS2:ETV1 translocation breakpoints. Top: genomic DNA extracted from LNCaP cells either non-transfected (E) or co-transfected with MeCP2 siRNA and FLAG-DOT1L expression vector (F and G) was subjected to PCR amplification using primers flanking Region II and I (E), Region III and I (F), or Region IV and I (G). Bottom: automated DNA sequencing aligned to ERG or ETV1 (green) and TMPRSS2 (orange) with genomic position of starting and ending nucleotides shown. Red box: common sequence shared by TMPRSS2 and ERG or ETV1. (H and I) Graphic illustration of translocation patterns corresponding to induced TMPRSS2:ERG (H) and TMPRSS2:ETV1 (I) translocations. Potential break/fusion sites are shown as Red oval: TMPRSS2:ERG; blue oval: TMPRSS2:ETV1; dot line: distinct fusion patterns.
Figure 4. AR-Dependent Local Chromatin Structural Alteration Sensitizes to Site-Specific Genotoxic Stress-Induced DSBs
(A-E) Top: schematic diagram showing the relative positions of break/fusion sites and potential AREs located on ERG and TMPRSS2 loci. Blue boxes, potential AREs; red boxes, break/fusion sites; black arrows, relative positions of PCR primers. Bottom: LNCaP cells were treated with DHT (10−7 M) (A), DHT (10−7 M) (B), IR (50 Gy) (C) or both (D) and DHT (10−7 M) (E) for time courses as indicated. ChIP analyses were performed with indicated antibodies on indicated regions (_n_=2, ±SEM). (F) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts with RPA2 siRNAs (_n_=3, ±SEM).
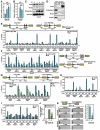
Figure 5. Mechanisms that Initiate Extended DNA Breaks in AR-Dependent Tumor Translocations
(A-C) Induction of AID expression by AR agonist and genotoxic stress. (_n_=3, ±SEM). (D) DHT-dependent interaction between AR and AID. Immunoprecipitates of anti-AR were subjected to immunoblotting analysis with indicated antibodies. Immunoblotting of E2F1 was included as negative control. (E) Ligand-dependent Myc-AID recruitment to AR-binding sites (_n_=2, ±SEM). (F and G) The recruitment of AID and Gadd45 to AR binding sites is mediated by liganded receptor. LNCaP cells were treated with ethanol (EtOH), DHT (10−7 M), or Bicalutamide (CDX, 10 μM) for 1hr followed by ChIP analyses with anti-Myc (F) or anti-AR and anti-Gadd45 (G) antibodies on indicated regions (_n_=2, ±SEM). (H and I) AID contributes to DSBs generation. ChIP analyses were performed on control siRNA or AID siRNA transfected, DHT+IR treated (4hr) LNCaP cells with anti-Ku80 antibodies (_n_=2, ±SEM) (H) or anti-BrdU antibody following BrdU labeling by TdT (I) on indicated regions. (J) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP cells transfected with AID siRNA (_n_=3, ±SEM). (K) Left: representative agarose gels with PCR products corresponding to TMPRESS2:ERGa translocations (as illustrated in Figure 3H band a). The genomic DNA of control or AID siRNA transfected LNCaP cells were subjected to PCR using primers flanking ligation site (red oval). Right: Statistical analysis (_n_=3, ±SEM).
Figure 6. Protective Effects of PIWIs on Chromosomal Translocation
(A) Left: summary of identified RGYW/WRCY motif related mutation. The fusion chromatins of ERG region A in control siRNA and AID siRNA samples or ERG region B were amplified as in Figure 3E. Right: statistics analysis (_n_=3, ±SEM). (B) Ligand-dependent UNG recruitment to intronic regions of TMPRSS2 and ERG loci. (C) Recruitment of UNG to translocation regions is AID dependent. D) Quantitation of induced TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP or PrEC cells. NPT: normal prostate tissue. (E) Statistical comparison of induced TMPRSS2:ERGb (left) and TMPRSS2:ETV1b (right) fusion transcripts LNCaP and PrEC cells (48 samples per group and _n_=3, ±SEM). (F) The relative expression level of PIWIL1 in LNCaP and PrEC cells. (G) The expression level of LINE-1 ORF2 was examined in PrEC cells transfected with indicated siRNAs. (H) PIWIL1 knockdown enhances γH2AX enrichment at intronic ERG and TMPRSS2 break/fusion sites. (I and J) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP cells electroporated with PIWIL1 siRNA (I) or indicated plasmids (J) (_n_=3, ±SEM). (K) Left: representative agarose gels with PCR products corresponding to TMPRESS2:ERGa translocations (as illustrated in Figure 3H band a). Right: Statistical analysis (_n_=3, ±SEM).
Figure 7. PIWI Regulated LINE-1 ORF2 Endonuclease Contributes to Chromosomal Translocations
(A) The relative expression level of LINE-1 ORF2 in LNCaP and PrEC cells. (B) IR-dependent induction of LINE-1 ORF2 expression (_n_=3, ±SEM). (C) Overexpression of LINE-1 ORF2 enhances Ku80 enrichment at intronic ERG and TMPRSS2 break/fusion sites. (D) Examination of TMPRSS2:ERGb and TMPRSS2:ETV1b fusion transcripts in LNCaP cells electroporated with indicated plasmids (_n_=3, ±SEM). (E and F) ORF2 contributes to DSBs generation independent of AID. ChIP analyses with anti-Ku80 (E) or anti-AID (F) antibodies were performed in LNCaP cells electroporated with indicated plasmids (_n_=2, ±SEM). (G) Recruitment of LINE-1 ORF2 to translocation regions. ChIP analyses with anti-FLAG followed by anti-ORF2 antibodies were performed in LNCaP cells electroporated with FLAG-ORF2 plasmid (_n_=2, ±SEM). (H) The recruitment of ORF2 is independent of AID. ChIP analyses with anti-FLAG antibodies were performed in LNCaP cells electroporated with AID siRNA and FLAG-ORF2 plasmid (_n_=2, ±SEM). (I) Schematic illustration of molecular mechanisms of nuclear receptor-dependent non-random chromosomal translocations.
Comment in
- The dangers of transcription.
Mathas S, Misteli T. Mathas S, et al. Cell. 2009 Dec 11;139(6):1047-9. doi: 10.1016/j.cell.2009.11.037. Cell. 2009. PMID: 20005797 Free PMC article.
Similar articles
- NKX3.1 Suppresses TMPRSS2-ERG Gene Rearrangement and Mediates Repair of Androgen Receptor-Induced DNA Damage.
Bowen C, Zheng T, Gelmann EP. Bowen C, et al. Cancer Res. 2015 Jul 1;75(13):2686-98. doi: 10.1158/0008-5472.CAN-14-3387. Epub 2015 May 14. Cancer Res. 2015. PMID: 25977336 Free PMC article. - TMPRSS2:ERG blocks neuroendocrine and luminal cell differentiation to maintain prostate cancer proliferation.
Mounir Z, Lin F, Lin VG, Korn JM, Yu Y, Valdez R, Aina OH, Buchwalter G, Jaffe AB, Korpal M, Zhu P, Brown M, Cardiff RD, Rocnik JL, Yang Y, Pagliarini R. Mounir Z, et al. Oncogene. 2015 Jul;34(29):3815-25. doi: 10.1038/onc.2014.308. Epub 2014 Sep 29. Oncogene. 2015. PMID: 25263440 - TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer.
Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. Hermans KG, et al. Cancer Res. 2006 Nov 15;66(22):10658-63. doi: 10.1158/0008-5472.CAN-06-1871. Cancer Res. 2006. PMID: 17108102 - Mechanisms that promote and suppress chromosomal translocations in lymphocytes.
Gostissa M, Alt FW, Chiarle R. Gostissa M, et al. Annu Rev Immunol. 2011;29:319-50. doi: 10.1146/annurev-immunol-031210-101329. Annu Rev Immunol. 2011. PMID: 21219174 Review. - Mechanisms of oncogenic chromosomal translocations.
Byrne M, Wray J, Reinert B, Wu Y, Nickoloff J, Lee SH, Hromas R, Williamson E. Byrne M, et al. Ann N Y Acad Sci. 2014 Mar;1310:89-97. doi: 10.1111/nyas.12370. Epub 2014 Feb 16. Ann N Y Acad Sci. 2014. PMID: 24528169 Review.
Cited by
- Large-scale functional organization of long-range chromatin interaction networks.
Sandhu KS, Li G, Poh HM, Quek YL, Sia YY, Peh SQ, Mulawadi FH, Lim J, Sikic M, Menghi F, Thalamuthu A, Sung WK, Ruan X, Fullwood MJ, Liu E, Csermely P, Ruan Y. Sandhu KS, et al. Cell Rep. 2012 Nov 29;2(5):1207-19. doi: 10.1016/j.celrep.2012.09.022. Epub 2012 Oct 25. Cell Rep. 2012. PMID: 23103170 Free PMC article. - Modeling of the human alveolar rhabdomyosarcoma Pax3-Foxo1 chromosome translocation in mouse myoblasts using CRISPR-Cas9 nuclease.
Lagutina IV, Valentine V, Picchione F, Harwood F, Valentine MB, Villarejo-Balcells B, Carvajal JJ, Grosveld GC. Lagutina IV, et al. PLoS Genet. 2015 Feb 6;11(2):e1004951. doi: 10.1371/journal.pgen.1004951. eCollection 2015. PLoS Genet. 2015. PMID: 25659124 Free PMC article. - RNA-Seq mapping and detection of gene fusions with a suffix array algorithm.
Sakarya O, Breu H, Radovich M, Chen Y, Wang YN, Barbacioru C, Utiramerur S, Whitley PP, Brockman JP, Vatta P, Zhang Z, Popescu L, Muller MW, Kudlingar V, Garg N, Li CY, Kong BS, Bodeau JP, Nutter RC, Gu J, Bramlett KS, Ichikawa JK, Hyland FC, Siddiqui AS. Sakarya O, et al. PLoS Comput Biol. 2012;8(4):e1002464. doi: 10.1371/journal.pcbi.1002464. Epub 2012 Apr 5. PLoS Comput Biol. 2012. PMID: 22496636 Free PMC article. - ETS rearrangements in prostate cancer.
Rubin MA. Rubin MA. Asian J Androl. 2012 May;14(3):393-9. doi: 10.1038/aja.2011.145. Epub 2012 Apr 16. Asian J Androl. 2012. PMID: 22504874 Free PMC article. Review. - Fusion genes and their discovery using high throughput sequencing.
Annala MJ, Parker BC, Zhang W, Nykter M. Annala MJ, et al. Cancer Lett. 2013 Nov 1;340(2):192-200. doi: 10.1016/j.canlet.2013.01.011. Epub 2013 Jan 29. Cancer Lett. 2013. PMID: 23376639 Free PMC article. Review.
References
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Science. Vol. 316. New York, NY: 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control; pp. 744–747. - PubMed
- Berkovich E, Monnat RJ, Jr., Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nature protocols. 2008;3:915–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS034934/NS/NINDS NIH HHS/United States
- R37 DK039949/DK/NIDDK NIH HHS/United States
- KG080247/PHS HHS/United States
- DK18477/DK/NIDDK NIH HHS/United States
- CA97134/CA/NCI NIH HHS/United States
- NS34934/NS/NINDS NIH HHS/United States
- R01 HL065445/HL/NHLBI NIH HHS/United States
- DK39949/DK/NIDDK NIH HHS/United States
- R01 CA097134/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials