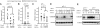The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation - PubMed (original) (raw)
The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation
Xuemei Tong et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Tumor cells are metabolically reprogrammed to fuel cell proliferation. Most transformed cells take up high levels of glucose and produce ATP through aerobic glycolysis. In cells exhibiting aerobic glycolysis, a significant fraction of glucose carbon is also directed into de novo lipogenesis and nucleotide biosynthesis. The glucose-responsive transcription factor carbohydrate responsive element binding protein (ChREBP) was previously shown to be important for redirecting glucose metabolism in support of lipogenesis in nonproliferating hepatocytes. However, whether it plays a more generalized role in reprogramming metabolism during cell proliferation has not been examined. Here, we demonstrated that the expression of ChREBP can be induced in response to mitogenic stimulation and that the induction of ChREBP is required for efficient cell proliferation. Suppression of ChREBP resulted in diminished aerobic glycolysis, de novo lipogenesis, and nucleotide biosynthesis, but stimulated mitochondrial respiration, suggesting a metabolic switch from aerobic glycolysis to oxidative phosphorylation. Cells in which ChREBP was suppressed by RNAi exhibited p53 activation and cell cycle arrest. In vivo, suppression of ChREBP led to a p53-dependent reduction in tumor growth. These results demonstrate that ChREBP plays a key role both in redirecting glucose metabolism to anabolic pathways and suppressing p53 activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
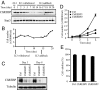
ChREBP protein level is regulated by growth factor signaling and is required for cell proliferation. Data in (A–C) are representative of at least three experiments. Data in (D) and (E) are presented as the mean ± SD of triplicate samples. (A) Western blot analysis of protein extracts harvested at the times indicated using antibodies to ChREBP and Stat3. The Stat3 blot serves as a loading control. IL-3-dependent bax−/−bak−/− mouse hematopoietic cells were grown in IL-3 and subjected to IL-3 withdrawal for 2 weeks, followed by restimulation with IL-3 at day 14. (B) Cell numbers of bax−/−bak−/− hematopoietic cultures that were grown in the presence or absence of IL-3 as described in A. (C) Western blot analysis of protein extracts of HCT116 cells transiently transfected with siRNA for control (Ctrl) and ChREBP (ChREBP1 and 2) using antibodies to ChREBP and tubulin at days 3 and 8 posttransfection. Human ChREBP protein displayed as a doublet whereas mouse ChREBP protein was a single band (A), which correlated with the presence of different ChREBP transcription variants in human. (D) Cell proliferation of HCT116 cells transfected with siRNA oligonucleotides for control (Ctrl) and ChREBP (ChREBP1 and 2). (E) Cell viability of HCT116 cells transfected with the indicated siRNA oligonucleotides at day 5 post-transfection.
Fig. 2.
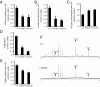
Suppression of ChREBP leads to reduced aerobic glycolysis and anabolic metabolism, accompanied by increased mitochondrial oxygen consumption in HCT116 cells. In (A–E), data are presented as the mean ± SD of triplicate samples. Data in (F) are representative of at least three experiments. (A) Glucose uptake and (B) lactate production of HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by cell number. *, P < 0.01. (C) Oxygen consumption of HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by cell number. *, P < 0.005. (D) Measurement of RNA synthesis from D-[U-14C6] glucose in HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by RNA amount. *, P < 0.0001. (E) Measurement of lipid produced from D-[6-14C] glucose in HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by cell number. *, P < 0.001. (F) Spectra of HCT116 cells transfected with control (Ctrl) and ChREBP2 siRNA and incubated with D-[1,6-13C2] glucose at day 3 post-transfection. Glut-2, Glut-4, and Lac-3 represent [2-13C] glutamate, [4-13C] glutamate and [3-13C] lactate, respectively.
Fig. 3.
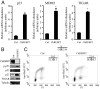
Attenuation of ChREBP activates p53 and induces cell cycle arrest. Data in (A) are presented as the mean ± SD of triplicate samples. Data in (B) and (C) are representative of at least three experiments. (A) Quantitative PCR analysis of p21, MDM2, and TIGAR in HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. *, P < 0.01. (B) Western blot analysis of HCT116 cells transfected with either control or ChREBP2 siRNA using indicated antibodies at day 3 post-transfection. (C) FACS analysis for BrdU incorporation and DNA content (PI) of HCT116 cells transfected with either control or ChREBP2 siRNA and pulse labeled with BrdU at day 3 post-transfection. Numbers indicate the percentage of cells in the G1, S, and G2/M phases.
Fig. 4.
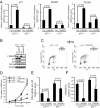
p53 is an important mediator of the growth and metabolic phenotype induced by ChREBP suppression. Data in (A) and (D–F) are presented as the mean ± SD of triplicate samples. Data in (B) and (C) are representative of at least three experiments. (A) Quantitative PCR analysis of p21, MDM2 and TIGAR in p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. (B) Western blot analysis of p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA using indicated antibodies at day 3 post-transfection. (C) FACS analysis for BrdU incorporation and DNA content (PI) of p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA and pulse labeled with BrdU at day 3 post-transfection. Numbers indicate the percentage of cells in the G1, S, and G2/M phases. (D) Cell proliferation of p53+/+ and p53−/− HCT116 cells transfected with siRNA for control and ChREBP2. (E) Glucose uptake of p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. The data were normalized by cell number. (F) Measurement of RNA synthesis from D-[U-14C6] glucose in p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. The data were normalized by RNA amount.
Fig. 5.
ChREBP suppression reduced tumor growth in vivo via a p53-dependent mechanism. (A–C) Charts depicting the mass of s.c. tumors formed in nude mice 18 days (A and B) and 10 days (C) after injection of indicated HCT116 stable transfectants. (A) Clones of p53+/+ HCT116 cells: ctrl-1 was generated by stable transfection of control pSM2C vector, while independent clones ChREBP-1 and ChREBP-12 were generated using the pSM2C-ChREBP shRNA plasmid. (B) Pools of p53+/+ HCT116 ChREBP-1 cells stably transfected with either empty pcDNA3 vector (Ctrl) or with a pcDNA3-mutant ChREBP plasmid. (C) Clones of p53−/− HCT116 cells: ctrl-5 was generated by stable transfection of control pSM2C vector, while independent clones ChREBP-17 and ChREBP-24 were generated using the pSM2C-ChREBP shRNA plasmid. (D and E) Western blot analysis of tumor protein extracts demonstrating efficient suppression of ChREBP levels in p53+/+ and p53−/− HCT116 cells stably transfected with ChREBP shRNA constructs as described in A–C. (D) Lanes 1–3 and 4–7 represent protein samples from two different animals, respectively. (E) Lanes 1–2, 3, and 4–7 represent protein samples from three different animals, respectively.
Similar articles
- Carbohydrate response element binding protein (ChREBP) correlates with colon cancer progression and contributes to cell proliferation.
Lei Y, Zhou S, Hu Q, Chen X, Gu J. Lei Y, et al. Sci Rep. 2020 Mar 6;10(1):4233. doi: 10.1038/s41598-020-60903-9. Sci Rep. 2020. PMID: 32144313 Free PMC article. - ChREBP, a glucose-responsive transcriptional factor, enhances glucose metabolism to support biosynthesis in human cytomegalovirus-infected cells.
Yu Y, Maguire TG, Alwine JC. Yu Y, et al. Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):1951-6. doi: 10.1073/pnas.1310779111. Epub 2014 Jan 21. Proc Natl Acad Sci U S A. 2014. PMID: 24449882 Free PMC article. - The ubiquitination ligase SMURF2 reduces aerobic glycolysis and colorectal cancer cell proliferation by promoting ChREBP ubiquitination and degradation.
Li Y, Yang D, Tian N, Zhang P, Zhu Y, Meng J, Feng M, Lu Y, Liu Q, Tong L, Hu L, Zhang L, Yang JY, Wu L, Tong X. Li Y, et al. J Biol Chem. 2019 Oct 4;294(40):14745-14756. doi: 10.1074/jbc.RA119.007508. Epub 2019 Aug 13. J Biol Chem. 2019. PMID: 31409643 Free PMC article. - ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome.
Iizuka K, Horikawa Y. Iizuka K, et al. Endocr J. 2008 Aug;55(4):617-24. doi: 10.1507/endocrj.k07e-110. Epub 2008 May 19. Endocr J. 2008. PMID: 18490833 Review. - The Role of Carbohydrate Response Element Binding Protein in Intestinal and Hepatic Fructose Metabolism.
Iizuka K. Iizuka K. Nutrients. 2017 Feb 22;9(2):181. doi: 10.3390/nu9020181. Nutrients. 2017. PMID: 28241431 Free PMC article. Review.
Cited by
- Genome-Wide Analysis of ChREBP Binding Sites on Male Mouse Liver and White Adipose Chromatin.
Poungvarin N, Chang B, Imamura M, Chen J, Moolsuwan K, Sae-Lee C, Li W, Chan L. Poungvarin N, et al. Endocrinology. 2015 Jun;156(6):1982-94. doi: 10.1210/en.2014-1666. Epub 2015 Mar 9. Endocrinology. 2015. PMID: 25751637 Free PMC article. - The Pentose Phosphate Pathway Dynamics in Cancer and Its Dependency on Intracellular pH.
Alfarouk KO, Ahmed SBM, Elliott RL, Benoit A, Alqahtani SS, Ibrahim ME, Bashir AHH, Alhoufie STS, Elhassan GO, Wales CC, Schwartz LH, Ali HS, Ahmed A, Forde PF, Devesa J, Cardone RA, Fais S, Harguindey S, Reshkin SJ. Alfarouk KO, et al. Metabolites. 2020 Jul 11;10(7):285. doi: 10.3390/metabo10070285. Metabolites. 2020. PMID: 32664469 Free PMC article. Review. - Transcriptomic analyses of gastrulation-stage mouse embryos with differential susceptibility to alcohol.
Boschen KE, Ptacek TS, Berginski ME, Simon JM, Parnell SE. Boschen KE, et al. Dis Model Mech. 2021 Jun 1;14(6):dmm049012. doi: 10.1242/dmm.049012. Epub 2021 Jun 17. Dis Model Mech. 2021. PMID: 34137816 Free PMC article. - The Roles of Carbohydrate Response Element Binding Protein in the Relationship between Carbohydrate Intake and Diseases.
Iizuka K. Iizuka K. Int J Mol Sci. 2021 Nov 8;22(21):12058. doi: 10.3390/ijms222112058. Int J Mol Sci. 2021. PMID: 34769488 Free PMC article. Review. - Myc, mondo, and metabolism.
Sloan EJ, Ayer DE. Sloan EJ, et al. Genes Cancer. 2010 Jun;1(6):587-96. doi: 10.1177/1947601910377489. Genes Cancer. 2010. PMID: 21113411 Free PMC article.
References
- Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. - PubMed
- Boros LG, et al. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. - PubMed
- Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. - PubMed
- Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature. 2008;452:230–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous