Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway - PubMed (original) (raw)
Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway
Yiyong Liu et al. J Biol Chem. 2010.
Abstract
At clinically relevant doses, chemotherapeutic S(N)1 DNA methylating agents induce an ATR-mediated checkpoint response in human cells that is dependent on functional MutSalpha and MutLalpha. Deficiency of either mismatch repair activity renders cells highly resistant to this class of drug, but the mechanisms linking mismatch repair to checkpoint activation have remained elusive. In this study we have systematically examined the interactions of human MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway using both nuclear extracts and purified proteins. Using nuclear co-immunoprecipitation, we have detected interaction of MutSalpha with ATR, TopBP1, Claspin, and Chk1 and interaction of MutLalpha with TopBP1 and Claspin. We were unable to detect interaction of MutSalpha or MutLalpha with Rad17, Rad9, or replication protein A in the extract system. Use of purified proteins confirmed direct interaction of MutSalpha with ATR, TopBP1, and Chk1 and of MutLalpha with TopBP1. MutSalpha-Claspin and MutLalpha-Claspin interactions were not demonstrable with purified proteins, suggesting that extract interactions are indirect or depend on post-translational modification. Use of a modified chromatin immunoprecipitation assay showed that proliferating cell nuclear antigen, ATR, TopBP1, and Chk1 are recruited to chromatin in a MutLalpha- and MutSalpha-dependent fashion after N-methyl-N'-nitro-N-nitrosoguanidine treatment. However, chromatin enrichment of replication protein A, Claspin, Rad17-RFC, and Rad9-Rad1-Hus1 was not detected in these experiments. Although our failure to observe enrichment of the latter activities could be due to sensitivity limitations, these observations may indicate a novel mechanism for ATR activation.
Figures
FIGURE 1.
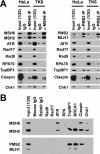
Reciprocal nuclear co-immunoprecipitation. A, immunoprecipitation from nuclear extracts of HeLa or TK6 cells was done using anti-MSH6 or anti-PMS2 antibodies as described under “Experimental Procedures.” The lanes marked as Input (1/20) indicate that 120 of total extract was loaded. Mouse IgG was used as a control for MSH6 or PMS2 antibody. The labels on the left indicate proteins co-precipitating with MSH6 or PMS2 as judged by Western blot after SDS-PAGE. Although some ATR signal is evident in the IgG control lane, quantitation of Western results demonstrated that the ATR signal in anti-MSH6 immunoprecipitates is 3–4-fold higher than that in the control sample. B, co-immunoprecipitation from nuclear extracts of TK6 cells was done by using antibodies against the checkpoint proteins indicated at the top of the figure. MutSα and MutLα in immunoprecipitates were evaluated after SDS-PAGE using antibodies directed against MSH2 and MSH6 (upper panel) or MLH1 and PMS2 (lower panel).
FIGURE 2.
Interactions of MutSα or MutLα with purified checkpoint proteins. A, MutSα or MutLα was incubated with FLAG-ATR as described under “Experimental Procedures.” Anti-FLAG M2 affinity gel was used to pull down FLAG-ATR and its interacting proteins. Incubation with FLAG peptide (20 n
m
) was used as a control for FLAG-ATR. ATR, MSH6, and MLH1 in the precipitates were determined by Western blot after SDS-PAGE. B, 9-1-1 was incubated alone, or with MutSα or MutLα followed by pulldown with anti-MSH2 or anti-PMS2 bound to protein G beads (“Experimental Procedures”). RAD9, MSH2, and PMS2 in the precipitates were determined by Western blot after SDS-PAGE. C, procedure was as in B, except RPA was used instead of 9-1-1, and RPA70 was probed in the Western blot. D, procedure was as in A except that glutathione-Sepharose beads were used to pull down GST-TopBP1 and its interacting partners. GST was used as a negative control for GST-TopBP1. E, procedure was as in D except GST-Chk1 was used instead of GST-TopBP1. F, after incubation with FLAG-Claspin, MutSα or MutLα was pulled down using anti-MSH2 or anti-MLH1 bound to protein G beads (right lanes). Negative controls included incubation of FLAG-Claspin with protein G beads alone or incubation of FLAG-Claspin with protein G beads and MSH2 or MLH1 antibody in the absence of MutSα or MutLα (lanes marked as α-MSH2 or α-MLH1). G, procedure was as in F except that incubation with Rad17-RFC was substituted for FLAG-Claspin.
FIGURE 3.
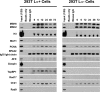
Modified ChIP assay using 293T Lα+ and 293T Lα− cells. Immunoprecipitation of micrococcal nuclease-digested formaldehyde-cross-linked chromatin samples with anti-MSH6 was performed as described under “Experimental Procedures.” After cross-link reversal, the samples were resolved by electrophoresis on SDS gels, which were probed by Western blot for proteins indicated on the left side of the figure. Negative controls included incubation of chromatin samples with protein G beads (Beads only) or incubation with protein G beads and mouse IgG (Mouse IgG). In each panel, the numbers above the right six lanes indicate the time after MNNG exposure in hours. Zero time samples were removed immediately prior to MMNG addition. Input (1/20) indicates that 5% of the zero time chromatin sample was loaded.
FIGURE 4.
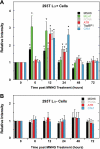
Quantification of protein bands for modified ChIP assay using 293T Lα+ and 293T Lα− cells. Band intensities of MSH6, histone H3, MLH1, PCNA, ATR, TopBP1, and Chk1 at 0–72 h post MNNG treatment were quantified using Image J (National Institutes of Health) and for each time sample normalized to the corresponding histone H3 signal. The relative intensity was then calculated by dividing the normalized intensity at each time by the zero time sample on the same gel. The results from triplicate experiments are expressed as the means ± S.D. The values that differ significantly from zero time relative intensities are indicated with asterisks (p < 0.05, two-sample t test). Quantification of results obtained with 293T Lα+ and 293T Lα− chromatin samples are shown in A and B, respectively.
FIGURE 5.
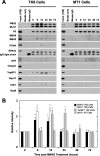
Modified ChIP assay using TK6 and MT1 cells. A, MLH1 antibody was used to immunoprecipitate MutLα. The assay was done as in Fig. 3. B, quantification of protein bands for the modified ChIP assay using TK6 and MT cells. The band intensity of MutLα and MSH2 at 0–72 h post MNNG treatment was quantified and analyzed as for Fig. 4.
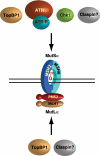
FIGURE 6.
MutSα and MutLα as a scaffold for recruitment of checkpoint proteins. Based on the results of this study and previous findings of others (29, 41, 49, 61), we suggest that the MutSα·MutLα complex with a DNA lesion may serve as a scaffold for recruitment of components of the ATR-Chk1 pathway.
Similar articles
- Interaction between Rad9-Hus1-Rad1 and TopBP1 activates ATR-ATRIP and promotes TopBP1 recruitment to sites of UV-damage.
Ohashi E, Takeishi Y, Ueda S, Tsurimoto T. Ohashi E, et al. DNA Repair (Amst). 2014 Sep;21:1-11. doi: 10.1016/j.dnarep.2014.05.001. Epub 2014 May 27. DNA Repair (Amst). 2014. PMID: 25091155 - Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation.
Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Liu S, et al. Mol Cell Biol. 2006 Aug;26(16):6056-64. doi: 10.1128/MCB.00492-06. Mol Cell Biol. 2006. PMID: 16880517 Free PMC article. - Regulation of ATR-CHK1 signaling by ubiquitination of CLASPIN.
Zhu X, Zheng XY, Gong P, Xu X. Zhu X, et al. Biochem Soc Trans. 2022 Oct 31;50(5):1471-1480. doi: 10.1042/BST20220729. Biochem Soc Trans. 2022. PMID: 36196914 Review.
Cited by
- Accidental Encounter of Repair Intermediates in Alkylated DNA May Lead to Double-Strand Breaks in Resting Cells.
Fujii S, Fuchs RP. Fujii S, et al. Int J Mol Sci. 2024 Jul 26;25(15):8192. doi: 10.3390/ijms25158192. Int J Mol Sci. 2024. PMID: 39125763 Free PMC article. Review. - FANCD2 counteracts O6-methylguanine-induced mismatch repair-dependent apoptosis.
Morita S, Fujikane R, Uechi Y, Matsuura T, Hidaka M. Morita S, et al. Mol Biol Rep. 2024 Jun 14;51(1):745. doi: 10.1007/s11033-024-09682-4. Mol Biol Rep. 2024. PMID: 38874758 - Systematic proximal mapping of the classical RAD51 paralogs unravel functionally and clinically relevant interactors for genome stability.
Simo Cheyou E, Boni J, Boulais J, Pinedo-Carpio E, Malina A, Sherill-Rofe D, Luo VM, Goncalves C, Bagci H, Maters A, Cuella-Martin R, Tabach Y, Del Rincon S, Côté JF, Rivera B, Orthwein A. Simo Cheyou E, et al. PLoS Genet. 2022 Nov 14;18(11):e1010495. doi: 10.1371/journal.pgen.1010495. eCollection 2022 Nov. PLoS Genet. 2022. PMID: 36374936 Free PMC article. - BK Polyomavirus Requires the Mismatch Repair Pathway for DNA Damage Response Activation.
Justice JL, Needham JM, Verhalen B, Jiang M, Thompson SR. Justice JL, et al. J Virol. 2022 Apr 27;96(8):e0202821. doi: 10.1128/jvi.02028-21. Epub 2022 Apr 7. J Virol. 2022. PMID: 35389233 Free PMC article. - Mismatch repair proteins play a role in ATR activation upon temozolomide treatment in MGMT-methylated glioblastoma.
Ganesa S, Sule A, Sundaram RK, Bindra RS. Ganesa S, et al. Sci Rep. 2022 Apr 6;12(1):5827. doi: 10.1038/s41598-022-09614-x. Sci Rep. 2022. PMID: 35388070 Free PMC article.
References
- Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) Chem. Rev. 106, 302–323 - PubMed
- Jiricny J. (2006) Nat. Rev. Mol. Cell Biol. 7, 335–346 - PubMed
- Li G. M. (2008) Cell Res. 18, 85–98 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM45190/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM032833/GM/NIGMS NIH HHS/United States
- GM32833/GM/NIGMS NIH HHS/United States
- R01 GM045190/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous