Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins - PubMed (original) (raw)
Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins
Ivar Ilves et al. Mol Cell. 2010.
Abstract
MCM2-7 proteins provide essential helicase functions in eukaryotes at chromosomal DNA replication forks. During the G1 phase of the cell cycle, they remain loaded on DNA but are inactive. We have used recombinant methods to show that the Drosophila MCM2-7 helicase is activated in complex with Cdc45 and the four GINS proteins (CMG complex). Biochemical activities of the MCM AAA+ motor are altered and enhanced through such associations: ATP hydrolysis rates are elevated by two orders of magnitude, helicase activity is robust on circular templates, and affinity for DNA substrates is improved. The GINS proteins contribute to DNA substrate affinity and bind specifically to the MCM4 subunit. All pairwise associations among GINS, MCMs, and Cdc45 were detected, but tight association takes place only in the CMG. The onset of DNA replication and unwinding may thus occur through allosteric changes in MCM2-7 affected by the association of these ancillary factors.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
Figure 1.. Reconstitution of CMG and MCM2-7 complexes.
(A) Purification protocol for the Drosophila CMG (B) Coomassie blue stained 10% SDS-PAGE gels of the purified MCM2-7 and CMG complexes (C) Stoichiometry of subunits in reconstituted MCM2-7 (dark columns) and CMG (white columns), based on laser densitometry scanning of SYPRO-red stained SDS-PAGE protein gels. Four independent MCM2-7 preparations and 3 CMG preparations were analyzed. The error bars in all the figures of this paper correspond to standard deviation, unless indicated otherwise. (D and E) Glycerol-gradient centrifugation analysis of the MCM2-7 and CMG. Western blotting with antibodies against MCM2, MCM4, and MCM5 subunits was used to detect protein in the fractions and quantification of these blots through densitometry using ImageJ software is shown in (D). Peak fractions were separated on 12% SDS-PAGE and silver stained (E).
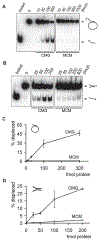
Figure 2.. DNA helicase assays with purified MCM2-7 and CMG.
(A and B) Autoradiographs of the reaction products separated on PAGE. Indicated amounts of CMG or MCM2-7 proteins in femtomoles were added to either a circular (A) or a forked substrate (B). Positions of double-stranded substrate and displaced oligo are indicated with arrows, ‘boiled’ and ‘0’ correspond to control lanes with heat denatured substrate or without protein, respectively. (C and D) Quantified results for two independent helicase assays with circular (C) or forked (D) DNA substrate. The graphs show percentage of displaced oligonucleotide as a function of protein added to reaction.
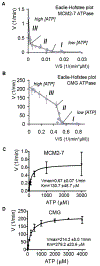
Figure 3.. Analysis of the ATP hydrolysis by MCM2-7 and CMG.
(A and B) Eadie-Hofstee plots of initial in vitro ATP hydrolysis kinetics for MCM2-7 and CMG (B). The regions that correspond to different kinetic modes at low (I), medium (II), or high (III) ATP concentrations are indicated. (C and D) Initial ATP hydrolysis rate kinetics for MCM2-7 (C) and CMG (D) plotted as a function of ATP concentration. Data were collected from 4 independent experiments, covering the entire tested ATP concentration range (0.15 μM – 4 mM for CMG, 0.03 μM – 3 mM for MCM2-7) with at least 3 overlapping datasets.
Figure 4.. Effect of ATPase site mutations in MCM subunits on ATPase and DNA helicase activity of CMG.
(A) Silver-stained 10% SDS-PAGE gels showing the individual protein complexes. ‘KA’ indicates a K to A substitution in the Walker A box of a given MCM subunit and ‘RA’ an R to A substitution in arginine finger domain, ‘wt’ indicates wild type CMG. (B) Vmax values (± standard error) for ATP hydrolysis reaction by wt CMG compared to mutant complexes. The values for MCM 2, 3, and 5 KA complexes are based on data from 5 independent series, for MCM 6 and 7 KA – from 2, and for MCM 4KA – from 1 series. (C and D) DNA helicase assays with wt or mutant CMG complexes. Autoradiographs of products separated on PAGE are shown in (C). Increasing amounts (25, 50, 100, or 200 fmol) of CMG complexes as indicated were added to the circular DNA substrate. In (D), the helicase activity of each mutant CMG complex is expressed relative to the wt as determined from the percent of oligo displaced by 100 or 200 fmol of protein (light grey and dark grey columns, respectively). The data for individual mutant complexes was collected from following number of independent series: 4KA – 9; 5KA - 5; 2KA, 6KA, 7KA, and 7RA – 4; 3KA, and 4KA/7RA – 2. (E) Graph illustrating the relative effects of individual Walker A box mutations around the MCM ring to the helicase activity of the CMG. The percent of activity retained after mutation is shown next to targeted ATPase sites, with 0-25% of wt indicated by ‘-‘ 25-50% - ‘+’, 50-75% - ‘++’, and 75-100% - ‘+++’.
Figure 5.. Interactions between the subunits of CMG.
(A - C) Western blots of the immunoprecipitated samples. Left panels show the probed blots with used antibodies indicated below, the right panels show the total protein staining of the same blots by colloidal gold. HA-tagged Sld5 subunit was used to precipitate the intact GINS complex and associated proteins in (A) as well as in (C); Cdc45 pull-down was performed in (B). CMG components in starting extract are shown on top (‘C’ – Cdc45; ‘M’ – MCM2-7; ‘G’ – GINS). (D) Silver-stained SDS-PAGE gel showing immunoaffinity pull-down of individual MCM subunits (as indicated on top) by GINS (HA-tag on Sld5). First four lines carry indicated amounts of MCM2-7 concentration marker. (E) Silver-stained SDS-PAGE gel showing the pull-down of individual MCM subunits by Cdc45. Arrowheads show the position of MCM2 and MCM4 bands. Western blotting analysis of the same samples is shown below. Each blot was probed with polyclonal antibodies detecting Cdc45 and individual MCMs; anti flag antibodies were used to probe flag-tagged MCM3 and MCM6. 100 ng of purified MCM2-7 (flag tag on MCM3) was loaded in each pair (‘M’) to compare different blots. (F) Interactions between the components of CMG complex based on data presented above. The thick line connecting GINS to MCM4 indicates a higher specificity than those indicated for the other pairs.
Figure 6.. MCM2-7 binds DNA more efficiently in complex with Cdc45 and GINS.
(A) Autoradiograph of the EMSA measuring DNA binding by MCM2-7 and CMG. Arrows show free forked probe and protein : DNA complexes. Increasing amounts (10, 30, 100, 300, and 1000 fmol) of MCM2-7 or CMG was added to the DNA substrate in the presence of ATP-γS. On all panels, ‘0’ corresponds to a control with no protein. (B) The quantification of data from (A), expressed as a percent of DNA probe that was shifted into a discreet slower moving band. The free probe signal from a control lane was set as 100% (C) EMSA showing that enhanced DNA biding by CMG is ATP dependent. ATP-γS was added as indicated to the binding reactions containing either 300 fmol CMG or 3 pmol of MCM2-7. (D) Alignment of C-terminal tails of the Psf3 protein in various organisms. (E) Silver-stained 12% SDS-PAGE gel of purified wt and CMG-delC complexes. Psf2 runs as a doublet because of an internal start codon (see Supplemental Experimental Procedures for details). (F and G) DNA binding by wt CMG and CMG-delC. Shown is an autoradiograph of EMSA with 10, 30, 100, and 300 fmol of proteins as indicated (F) and the quantification of data from three independent experiments with two different CMG-delC protein preparations (G). The activity was calculated as in (B) and expressed relative to wt activity in the same series for reactions with 100 fmol (dark grey columns) or 300 fmol (light grey) of protein. (H) Quantified results of DNA helicase assays with wt and CMG-delC complexes. Two different protein preparations were tested in duplicates on circular or forked substrates as indicated. Columns show the fraction of substrate displaced relative to wt activity by either 100 fmol or 200 fmol of protein (dark grey and light grey columns, respectively). (I) Coomassie-stained 12% SDS-PAGE gel of purified wt and GINS-delC complexes. (J) EMSA of DNA binding by increasing amounts (0.3, 1, 3, and 8 pmol per reaction) of wt GINS and GINS-delC.
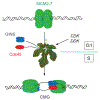
Figure 7.. Activation of the MCM helicase requires binding of GINS and Cdc45.
Double hexamers of MCM2-7 are loaded to DNA in G1 and the best available data suggests that these MCM rings are probably on pathway for DNA replication. The subsequent events require CDK regulated formation of potential chaperone complexes such as those associated with Dpb11 (See Botchan, 2007, for review) and the DDK modification of the amino-terminal tails of the MCMs. In the model discussed, these events lead to: A) release of the double hexamers to enable binding of activating GINS and Cdc45 proteins, and B) coupled to the binding of the activators, the MCMs isomerize so that DNA strands are organized to create a bidirectional replication fork at the time of initiation.
Similar articles
- DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome.
Costa A, Renault L, Swuec P, Petojevic T, Pesavento JJ, Ilves I, MacLellan-Gibson K, Fleck RA, Botchan MR, Berger JM. Costa A, et al. Elife. 2014 Aug 12;3:e03273. doi: 10.7554/eLife.03273. Elife. 2014. PMID: 25117490 Free PMC article. - Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase.
Moyer SE, Lewis PW, Botchan MR. Moyer SE, et al. Proc Natl Acad Sci U S A. 2006 Jul 5;103(27):10236-10241. doi: 10.1073/pnas.0602400103. Epub 2006 Jun 23. Proc Natl Acad Sci U S A. 2006. PMID: 16798881 Free PMC article. - The structural basis for MCM2-7 helicase activation by GINS and Cdc45.
Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. Costa A, et al. Nat Struct Mol Biol. 2011 Apr;18(4):471-7. doi: 10.1038/nsmb.2004. Epub 2011 Mar 6. Nat Struct Mol Biol. 2011. PMID: 21378962 Free PMC article. - The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism.
Li H, O'Donnell ME. Li H, et al. Bioessays. 2018 Mar;40(3):10.1002/bies.201700208. doi: 10.1002/bies.201700208. Epub 2018 Feb 6. Bioessays. 2018. PMID: 29405332 Free PMC article. Review. - Activation of the replicative DNA helicase: breaking up is hard to do.
Boos D, Frigola J, Diffley JF. Boos D, et al. Curr Opin Cell Biol. 2012 Jun;24(3):423-30. doi: 10.1016/j.ceb.2012.01.011. Epub 2012 Mar 16. Curr Opin Cell Biol. 2012. PMID: 22424671 Review.
Cited by
- Transcriptional activation is a conserved feature of the early embryonic factor Zelda that requires a cluster of four zinc fingers for DNA binding and a low-complexity activation domain.
Hamm DC, Bondra ER, Harrison MM. Hamm DC, et al. J Biol Chem. 2015 Feb 6;290(6):3508-18. doi: 10.1074/jbc.M114.602292. Epub 2014 Dec 23. J Biol Chem. 2015. PMID: 25538246 Free PMC article. - Interplay between Fanconi anemia and homologous recombination pathways in genome integrity.
Michl J, Zimmer J, Tarsounas M. Michl J, et al. EMBO J. 2016 May 2;35(9):909-23. doi: 10.15252/embj.201693860. Epub 2016 Apr 1. EMBO J. 2016. PMID: 27037238 Free PMC article. Review. - Termination of DNA replication forks: "Breaking up is hard to do".
Bailey R, Priego Moreno S, Gambus A. Bailey R, et al. Nucleus. 2015;6(3):187-96. doi: 10.1080/19491034.2015.1035843. Epub 2015 Apr 2. Nucleus. 2015. PMID: 25835602 Free PMC article. - Behavior of replication origins in Eukaryota - spatio-temporal dynamics of licensing and firing.
Musiałek MW, Rybaczek D. Musiałek MW, et al. Cell Cycle. 2015;14(14):2251-64. doi: 10.1080/15384101.2015.1056421. Epub 2015 Jun 1. Cell Cycle. 2015. PMID: 26030591 Free PMC article. Review. - Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication.
Li Y, Araki H. Li Y, et al. Genes Cells. 2013 Apr;18(4):266-77. doi: 10.1111/gtc.12040. Epub 2013 Mar 5. Genes Cells. 2013. PMID: 23461534 Free PMC article. Review.
References
- Aparicio OM, Weinstein DM, and Bell SP (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69. - PubMed
- Bell SP, and Dutta A (2002). DNA replication in eukaryotic cells. Annual Review of Biochemistry 71, 333–374. - PubMed
- Bochman ML, and Schwacha A (2007). Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and 5 define a slow ATP-dependent step. J. Biol. Chem 282, 33795–33804. - PubMed
- Bochman ML, and Schwacha A (2008). The MCM2-7 complex has in vitro helicase activity. Mol. Cell 31, 287–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous