Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF - PubMed (original) (raw)
Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF
Sonja J Heidorn et al. Cell. 2010.
Abstract
We describe a mechanism of tumorigenesis mediated by kinase-dead BRAF in the presence of oncogenic RAS. We show that drugs that selectively inhibit BRAF drive RAS-dependent BRAF binding to CRAF, CRAF activation, and MEK-ERK signaling. This does not occur when oncogenic BRAF is inhibited, demonstrating that BRAF inhibition per se does not drive pathway activation; it only occurs when BRAF is inhibited in the presence of oncogenic RAS. Kinase-dead BRAF mimics the effects of the BRAF-selective drugs and kinase-dead Braf and oncogenic Ras cooperate to induce melanoma in mice. Our data reveal another paradigm of BRAF-mediated signaling that promotes tumor progression. They highlight the importance of understanding pathway signaling in clinical practice and of genotyping tumors prior to administering BRAF-selective drugs, to identify patients who are likely to respond and also to identify patients who may experience adverse effects.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
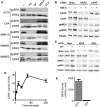
BRAF Inhibitors Activate CRAF, MEK, and ERK in RAS Mutant Cell Lines (A) A375, D04, MM415, MM485, and WM852 cells were treated with DMSO (−), PD184352 (PD; 1 μM), sorafenib (SF; 10 μM), 885-A (1 μM) and PLX4720 (PLX; 0.3 μM) for 4 hr. Cell extracts were western blotted for phospho-ERK (ppERK) and total ERK2 (loading control). (B and C) D04 cells were transfected with siRNA against NRAS or CRAF, or control (Mock) as indicated. After 48 hr the cells were treated with DMSO (−), 885-A (1 μM) or PLX4720 (PLX; 0.3 μM) for 4 hr. Cell lysates were western blotted for NRAS, CRAF, phospho-MEK (ppMEK), phospho-ERK (ppERK) and tubulin (loading control). (D) D04 cells were treated with 885-A for various times and endogenous CRAF kinase activity was measured. Data show fold activation of experimental triplicates compared to untreated cells with error bars to represent standard deviations from the means. (E) Endogenous BRAF kinase activity was measured in A375 or D04 cells. The results (arbitrary units per μg of cell protein) are the mean of an assay performed in triplicate with error bars to represent standard deviation from the mean.
Figure 2
BRAF Inhibitors Induce CRAF Binding to BRAF (A) WM852, D04, MM415 and MM485 cells were treated with DMSO (−), PD184352 (PD; 1 μM), sorafenib (SF; 10 μM), 885-A (1 μM) or PLX4720 (PLX; 0.3 μM) for 4 hr. Endogenous BRAF (IP: BRAF) or endogenous CRAF (IP: CRAF) were immunoprecipitated and the immunocomplexes were western blotted (WB) for BRAF or CRAF. BRAF, and CRAF levels in the cell lysates are also shown. (B) D04 cells were treated with DMSO (−), PD184352 (PD; 1 μM), sorafenib (SF; 10 μM) and PLX4720 (PLX; 0.3μM) for 4 hr. Endogenous CRAF (IP: CRAF) was immunoprecipitated and the immunocomplexes were western blotted (WB) for BRAF or CRAF. BRAF and CRAF levels in the cell lysates are shown. (C) SW620, HCT116 and WM1791c cells were treated with DMSO (−), PD184352 (PD; 1 μM), sorafenib (SF; 10 μM) or 885-A (1 μM) for 4 hr. Endogenous BRAF (IP: BRAF) or endogenous CRAF (IP: CRAF) were immunoprecipitated and the immunocomplexes were western blotted (WB) for BRAF or CRAF. The cell lysates were also blotted for BRAF, CRAF, phospho-ERK (ppERK) and total ERK2 (loading control). (D) A375 cells were treated with DMSO (−), PD184352 (PD; 1 μM), sorafenib (SF; 10 μM), 885-A (1 μM) or PLX4720 (PLX; 0.3 μM) for 4 hr. CRAF (IP: CRAF) was immunoprecipitated and the immunocomplexes were western blotted (WB) for BRAF or CRAF. BRAF and CRAF levels in the cell lysates are shown.
Figure 3
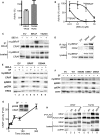
BRAF Binding to CRAF Requires RAS (A) Myc-epitope tagged CRAF or R89LCRAF (R89L), or an empty vector control (EV) were transfected into D04 cells. After 48 hr, the cells were treated with DMSO (−) or 885-A (1 μM) for 4 hr. Myc-tagged CRAF was immunoprecipitated (IP) and the immunocomplexes were western blotted (WB) for endogenous BRAF or myc-CRAF. Endogenous BRAF and myc-CRAF levels in the cell lysates are also shown. (B) Myc-epitope tagged BRAF or R188LBRAF (R188L) or an empty vector control (EV) were transfected into D04 cells. After 48 hr the cells were treated with DMSO (−) or 885-A (1 μM) for 4 hr. Myc-tagged BRAF was immunoprecipitated (IP) and the immunocomplexes were western blotted (WB) for myc-BRAF or endogenous CRAF. Myc-BRAF and endogenous-CRAF levels in the cell lysates are also shown. (C) Membrane or cytosol fractions were prepared from untreated (−) or 885-A (1 μM) treated D04 cells. BRAF, CRAF, Tubulin (cytosol control) and HRAS (membrane control) were western blotted in the total lysate (TL), cytosolic fraction (CYT) and membrane fraction (MEM). The graph shows the quantification of the relative levels of BRAF and CRAF in the membrane and cytosol fractions. (D) PMWK cells were pretreated with DMSO or 885-A (1 μM, 60 min) and then treated with EGF (10 ng/ml) for the times shown in minutes (min). Endogenous CRAF was immunoprecipitated (IP) and the precipitates were western blotted (WB) for BRAF and CRAF. The lysates were also western blotted for BRAF, CRAF, phospho-MEK (ppMEK), phospho-ERK (ppERK) and total ERK2. (E) D04 cells were treated with DMSO (−) or sorafenib (+; 10 μM) for 4 hr. Endogenous BRAF was immunoprecipitated and the immunocomplexes left untreated or incubated with calf intestinal phosphatase (CIP; 5U, 30°C, 30 min) in the presence or absence of phosphatase inhibitors (P'ase Inh). Immunocomplexes were western blotted for BRAF and CRAF. (F) D04 cells were treated with DMSO (−), PD184352 (PD; 1 μM) or 885-A (1 μM) for 4 hr. Endogenous CRAF (IP: CRAF) was immunoprecipitated and the immunocomplexes were western blotted (WB) for BRAF or CRAF. BRAF, CRAF, and phospho-ERK (ppERK) levels in the cell lysates are shown.
Figure 4
BRAF and Not CRAF Inhibition Drives CRAF Binding to BRAF and CRAF Activation (A) COS cells were transiently transfected with myc-epitope tagged BRAF, or T529NBRAF (T529N) in the presence of G12VHRAS (RAS) and their kinase activity was measured. The data represent one assay performed in triplicate, with error bars to represent standard deviations from the mean. Activity (%) is relative to wild-type BRAF activated by G12VHRAS. (B) As in (A) but immunocomplexes were treated with DMSO (−) or 885-A for 10 min prior to measuring their kinase activity. The data represent one assay performed in triplicate, with error bars to represent standard deviations from the mean. Activity (% control) is relative to the untreated kinase. (C) Myc-epitope tagged BRAF, T529NBRAF (T529N), or an empty vector control (EV) were transfected into D04 cells. After 48 hr the cells were treated with DMSO (−) or 885-A (1 μM) for 4 hr. The endogenous CRAF was immunoprecipitated and the immunocomplexes were western blotted for myc-BRAF or endogenous CRAF. Myc-BRAF and endogenous CRAF levels in the cell lysates are also shown. (D) Myc-epitope tagged BRAF, D594ABRAF (D594A), or an empty vector control (EV) were transfected into D04 cells. After 48 hr the myc-BRAF was immunoprecipitated (IP) and the immunocomplexes were western blotted for mycBRAF and endogenous CRAF. Myc-BRAF and endogenous CRAF levels in the cell lysates are also shown. (E) Myc-epitope tagged BRAF, D594ABRAF (D594A), or an empty vector control (EV) were transfected into D04 cells. After 48 hr the cells were treated with DMSO (−), sorafenib (SF; 10 μM) or 885-A (1 μM) for 4 hr. The cells extracts were western blotted for myc-BRAF, phospho-MEK (ppMEK), phospho-ERK and total ERK2 (loading control). Note that ERK2 runs as a doublet due to the separation of the phosphorylated and nonphosphorylated protein. (F) Myc-epitope tagged BRAF, K483MBRAF (K483M), D594VBRAF (D594V), D594ABRAF (D594A), or an empty vector control (EV) were transfected into D04 cells. After 48 hr, the cells were treated with DMSO (−) or sorafenib (SF; 10 μM) for 4 hr. Cell extracts were blotted for myc-BRAF, phospho-MEK (ppMEK), phospho-ERK (ppERK) and CRAF (loading control). (G) D04 cells were treated with sorafenib (10 μM) for various times and CRAF kinase activity was measured. Data is for one assay performed in triplicate, with error bars to represent standard deviations from the means. Inset: D04 cells were treated with sorafenib (SF) for 4 hr and CRAF was immunoprecipitated and western blotted for S338 phosphorylation (pS338). CRAF levels in the lysate are shown as a loading control. (H) D04 cells stably expressing flag-epitope tagged CRAF (CRAF) or T421NCRAF (T421N) were treated with DMSO (−), PD184352 (PD; 1 μM), sorafenib (SF; 10 μM) or 885-A (1 μM) for 4 hr. The flag-CRAF was immunoprecipitated (IP) and the immunocomplexes were western blotted for endogenous-BRAF or flag-CRAF. Endogenous BRAF, flag-CRAF and phosphorylated ERK (ppERK) levels in the cell lysates are also shown.
Figure 5
Oncogenic Kras and Kinase-Dead Braf Cooperate to Drive Tumorigenesis (A) Diagrammatic representation of targeted conditional BrafLSL-D594A allele used for D594ABraf expression in mouse melanocytes. The endogenous mouse Braf gene from exons 14–15 is represented. Exon 15 is mutated to express D594ABraf (15∗). LoxP sites are represented by triangles. The relative position of the wild-type BRAF minigene (MG) comprising exons 15–18 of BRAF, the transcription terminator (term) and the NeoR cassette are shown. Cre-recombinase mediated removal of these regions results in BrafLox-D594A, allowing expression of D594ABraf. (B) Photographs of the tails of tamoxifen-treated wild-type (WTKras/WTBraf), Kras+/LSL-G12D;Tyr::CreERT2+/o (G12DKras/WTBraf), or Kras+/LSL-G12D;Braf+/LSL-D594A;Tyr::CreERT2+/o (G12DKras/D594ABraf) mice. (C) Kaplan-Meier plots showing disease free progression of study mice. The controls consisted of 12 tamoxifen-treated Tyr::CreERT2+/o mice; 10 ethanol-treated Braf+/LSL-D594A;Tyr::CreERT2+/o mice and 6 ethanol-treated Kras+/LSL-G12D; Tyr::CreERT2+/o mice. The experimental groups consisted of 12 tamoxifen-treated Kras+/LSL-G12D;Tyr::CreERT2+/o (G12DKras), 24 tamoxifen-treated Braf+/LSL-D594A;Tyr::CreERT2+/o (D594ABraf) mice, and 3 tamoxifen-treated Kras+/LSL-G12D;Braf+/LSL-D594A;Tyr::CreERT2+/o (G12DKras/D594ABraf) mice. (D) Photographs of the feet of tamoxifen-treated wild-type (WTKras/WTBraf), or Kras+/LSL-G12D;Braf+/LSL-D594A;Tyr::CreERT2+/o (G12DKras/D594ABraf) mice. (E) Photograph showing a large tumor on the back of a tamoxifen-treated Kras+/LSL-G12D;Braf+/LSL-D594A;Tyr::CreERT2+/o (G12DKras/D594ABraf) mouse. The fur was removed to reveal the lesion. (F) Photomicrograph of a tumor from the back of a G12DKras/D594ABraf mouse. An area of ulceration is highlighted by the arrow. (G) High magnification photomicrograph of a section of tumor showing atypical cells, conspicuous nucleoli (arrowheads) and nuclear pseudo-inclusions (arrows). (H) High magnification photomicrograph of a section of tumor showing mitotic figures (arrows). (I) Photomicrograph of a section of tumor subjected to immunohistochemical analysis with antibodies against Ki67 (MIB1).
Figure 6
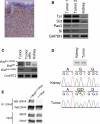
Tumors Induced by Kinase-Dead Braf and Oncogenic Kras Are Melanoma (A) Photomicrograph of a section of tumor subjected to immunohistochemical analysis with antibodies against S100. (B) RT-PCR analysis revealing expression of tyrosinase (Tyr), Dct, Pax3 and silver/gp100 (Si) in two independent tumors and kidney (control). GAPDH is used as a loading control. (C) PCR-mediated genotyping for wild-type Braf (BrafWT), Braf+/LSL-D594A and Tyr::CreERT2+/o alleles from a tumor sample, cells derived from the tumor and from kidney as a control. (D) PCR amplified fragment for Kras from kidney and tumor samples. Shown below is the sequencing trace for codons 11–13, together with the DNA and protein sequence (single amino acid code). (E) Endogenous CRAF was immunoprecipitated (IP) from cells derived from a tumor from a G12DKras/D594ABraf mouse (Kras+/LSL-G12D;Braf+/LSL-D594A;Tyr::CreERT2+/o), or from a tumor from a G12VKras mouse (β-actin+/LSL-G12VKras;Tyr::CreERT2+/o). The immunocomplexes were western blotted (WB) for Braf and Craf, and the levels of Braf and Craf in the cell lysates are also shown.
Figure 7
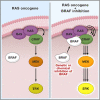
A Model of Paradoxical CRAF Activation by BRAF (A) In the presence of oncogenic RAS, BRAF is cytosolic, where it maintains itself in an inactive conformation in a manner that depends on its own kinase activity. CRAF is recruited to the plasma membrane by RAS and activates the pathway. (B) When BRAF is inhibited by genetic or chemical means, it is no longer autoinhibited and is recruited to the plasma membrane by RAS, where it binds to CRAF. Although BRAF does not itself signal, it can act as a scaffold to enhance CRAF activity and consequently enhance signaling through the pathway. (C) Pan-RAF inhibitors hyperactivate CRAF because they inhibit BRAF, but they simultaneously inhibit CRAF, leading to paradoxical activation of CRAF without pathway activation. (D) T421NCRAF (T421N) escapes the paradoxical activation by the pan-RAF inhibitors, because it no longer allows them to bind, so is freely activated due to BRAF inhibition.
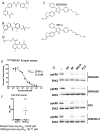
Figure S1
Characterization of 885-A, an Analog of SB590885, Relates to Figure 1 (A) Sorafenib, a pan-RAF, multi-kinase class II inhibitor. (B) PLX4720, a selective class I BRAF inhibitor. (C) SB590885 and its analog 885-A, selective class I BRAF inhibitors. (D) PD184352, a MEK inhibitor (CI1040). (E) Inhibition of V600EBRAF by 885-A in vitro. Insect cell purified V600EBRAF kinase activity was measured at increasing concentrations of 885-A using a 96-well DELFIA-based assay system (Perkin Elmer, Amersham, UK). The assays were performed in the linear range of the assay and in duplicate using a concentration response of 11 points. IC50 values were determined using GraphPad Prism software (GraphPad Software, San Diego, CA) and the reported IC50 values are the mean of 3 independent assays. (F) 885-A selectively inhibits the growth of BRAF mutant melanoma cells. The growth inhibitory activity of 885-A against a panel of cell lines (7 V600 BRAF mutant lines; 10 wild-type BRAF lines) is represented. The individual IC50 values for each line are represented by the symbols, and the horizontal line represents the mean IC50 for these two populations. (G) SKMel24, SKMel28, D25 and WM266.4 cells were treated with DMSO (-), PD184352 (PD; 1μM), sorafenib (SF; 10 μM), 885-A (1μM) and PLX4720 (PLX; 0.3μM) for 4 hr. Cell extracts were western blotted for phospho-ERK (ppERK) and total ERK2 (loading control).
Figure S2
MEK, pan-RAF or BRAF Inhibitors Do Not Induce Strong BRAF Binding to CRAF in BRAF Mutant Melanoma Cell Lines, Relates to Figure 2 D25 and WM266.4 cells were treated with DMSO (-), PD184352 (PD; 1μM), sorafenib (SF; 10 μM), 885-A (1μM) or PLX4720 (PLX; 0.3μM) for 4 hr. Endogenous CRAF (IP: CRAF) was immunoprecipitated and the immunocomplexes were western blotted (WB) for BRAF and CRAF. BRAF, CRAF, phospho-MEK (ppMEK), phospho-ERK (ppERK) and total ERK2 levels in the cell lysates are also shown.
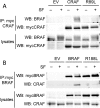
Figure S3
BRAF Binding to CRAF Requires BRAF and CRAF Binding to RAS, Relates to Figure 3 (A) Myc-epitope tagged CRAF or R89LCRAF (R89L), or an empty vector control (EV) were transfected into D04 cells. After 48 hr the cells were treated with DMSO (-) or sorafenib (SF, +; 10 μM) for 4 hr. The myc-CRAF was immunoprecipitated (IP) and the immunocomplexes were western blotted (WB) for endogenous BRAF or myc-CRAF. Endogenous BRAF and myc-CRAF levels in the cell lysates are also shown. (B) Myc-epitope tagged BRAF or R188LBRAF (R188L) or an empty vector control (EV) were transfected into D04 cells. After 48 hr the cells were treated with DMSO (-) or sorafenib (SF, +; 10 μM) for 4 hr. The myc-BRAF was immunoprecipitated (IP) and the immunocomplexes were western blotted (WB) for myc-BRAF or endogenous CRAF. Myc-BRAF and endogenous-CRAF levels in the cell lysates are also shown.
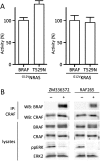
Figure S4
Characterization of T529NBRAF and the pan-RAF Inhibitors ZM336372 and RAF265, Relates to Figure 4 (A) T529NBRAF is activated by NRAS and KRAS. COS cells were transiently transfected with myc-epitope tagged BRAF, or T529NBRAF (T529N) in the presence of G12VNRAS or G12VKRAS as indicated. BRAF kinase activity was measured in an immunoprecipitation kinase assay. The data represent one assay performed in triplicate, with error bars to represent standard deviations from the mean. Activity (%) is relative to wild-type BRAF activated by G12VNRAS or G12VKRAS as appropriate. (B) Pan-RAF inhibitors drive BRAF binding to CRAF but not ERK activation. DO4 cells were treated with ZM336372 or RAF265 for 4 hr. CRAF (IP: CRAF) was immunoprecipitated and the immnocomplexes were western blotted (WB) for BRAF or CRAF. BRAF and CRAF levels in the cell lysates are shown and the lysates were also western blotted for phosphorylated ERK (ppERK) and total ERK2.
Comment in
- The brothers RAF.
Kwong LN, Chin L. Kwong LN, et al. Cell. 2010 Jan 22;140(2):180-2. doi: 10.1016/j.cell.2010.01.013. Cell. 2010. PMID: 20141832 - Anticancer drugs: RAF complexities spark caution.
McCarthy N. McCarthy N. Nat Rev Drug Discov. 2010 Apr;9(4):271. doi: 10.1038/nrd3153. Nat Rev Drug Discov. 2010. PMID: 20535844 No abstract available.
Similar articles
- Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation.
Su F, Bradley WD, Wang Q, Yang H, Xu L, Higgins B, Kolinsky K, Packman K, Kim MJ, Trunzer K, Lee RJ, Schostack K, Carter J, Albert T, Germer S, Rosinski J, Martin M, Simcox ME, Lestini B, Heimbrook D, Bollag G. Su F, et al. Cancer Res. 2012 Feb 15;72(4):969-78. doi: 10.1158/0008-5472.CAN-11-1875. Epub 2011 Dec 28. Cancer Res. 2012. PMID: 22205714 - Registered report: Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF.
Bhargava A, Anant M, Mack H; Reproducibility Project: Cancer Biology; Reproducibility Project Cancer Biology. Bhargava A, et al. Elife. 2016 Feb 17;5:e11999. doi: 10.7554/eLife.11999. Elife. 2016. PMID: 26885666 Free PMC article. - RAF proteins exert both specific and compensatory functions during tumour progression of NRAS-driven melanoma.
Dorard C, Estrada C, Barbotin C, Larcher M, Garancher A, Leloup J, Beermann F, Baccarini M, Pouponnot C, Larue L, Eychène A, Druillennec S. Dorard C, et al. Nat Commun. 2017 May 12;8:15262. doi: 10.1038/ncomms15262. Nat Commun. 2017. PMID: 28497782 Free PMC article. - Raf kinases in cancer-roles and therapeutic opportunities.
Maurer G, Tarkowski B, Baccarini M. Maurer G, et al. Oncogene. 2011 Aug 11;30(32):3477-88. doi: 10.1038/onc.2011.160. Epub 2011 May 16. Oncogene. 2011. PMID: 21577205 Review. - MEK and RAF inhibitors for BRAF-mutated cancers.
Belden S, Flaherty KT. Belden S, et al. Expert Rev Mol Med. 2012 Oct 12;14:e17. doi: 10.1017/erm.2012.11. Expert Rev Mol Med. 2012. PMID: 23058743 Review.
Cited by
- RAS signaling in carcinogenesis, cancer therapy and resistance mechanisms.
Yang X, Wu H. Yang X, et al. J Hematol Oncol. 2024 Nov 9;17(1):108. doi: 10.1186/s13045-024-01631-9. J Hematol Oncol. 2024. PMID: 39522047 Free PMC article. Review. - Therapeutic vulnerabilities and pan-cancer landscape of BRAF class III mutations in epithelial solid tumors.
Özgü E, Kaplan BG, Sivakumar S, Sokol ES, Aydın E, Tokat ÜM, Adibi A, Karakoç EG, Hu J, Kurzrock R, Demiray M. Özgü E, et al. BJC Rep. 2024 Oct 8;2(1):77. doi: 10.1038/s44276-024-00086-2. BJC Rep. 2024. PMID: 39516363 Free PMC article. - The polymeric fluoropyrimidine CF10 overcomes limitations of 5-FU in pancreatic ductal adenocarcinoma cells through increased replication stress.
Finan JM, Di Niro R, Park SY, Jeong KJ, Hedberg MD, Smith A, McCarthy GA, Haber AO, Muschler J, Sears RC, Mills GB, Gmeiner WH, Brody JR. Finan JM, et al. Cancer Biol Ther. 2024 Dec 31;25(1):2421584. doi: 10.1080/15384047.2024.2421584. Epub 2024 Nov 8. Cancer Biol Ther. 2024. PMID: 39513592 Free PMC article. - Navigating the changing landscape of BTK-targeted therapies for B cell lymphomas and chronic lymphocytic leukaemia.
Stanchina MD, Montoya S, Danilov AV, Castillo JJ, Alencar AJ, Chavez JC, Cheah CY, Chiattone C, Wang Y, Thompson M, Ghia P, Taylor J, Alderuccio JP. Stanchina MD, et al. Nat Rev Clin Oncol. 2024 Nov 1. doi: 10.1038/s41571-024-00956-1. Online ahead of print. Nat Rev Clin Oncol. 2024. PMID: 39487228 Review. - Atypical Exon 2/3 Mutants G48C, Q43K, and E37K Present Oncogenic Phenotypes Distinct from Characterized NRAS Variants.
Fran MAG, Leaño DMG, de Borja JAD, Uy CJT, Andresan AAR, Sacdalan DL, Garcia RL. Fran MAG, et al. Cells. 2024 Oct 12;13(20):1691. doi: 10.3390/cells13201691. Cells. 2024. PMID: 39451209 Free PMC article.
References
- Cameron A.J., Escribano C., Saurin A.T., Kostelecky B., Parker P.J. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat. Struct. Mol. Biol. 2009;16:624–631. - PubMed
- Dhomen N., Reis-Filho J.S., da Rocha Dias S., Hayward R., Savage K., Delmas V., Larue L., Pritchard C., Marais R. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell. 2009;15:294–303. - PubMed
- Dumaz N., Hayward R., Martin J., Ogilvie L., Hedley D., Curtin J.A., Bastian B.C., Springer C., Marais R. In Melanoma, RAS Mutations Are Accompanied by Switching Signaling from BRAF to CRAF and Disrupted Cyclic AMP Signaling. Cancer Res. 2006;66:9483–9491. - PubMed
Supplemental References
- Monks, A., Scudiero, D., Skehan, P., Shoemaker, R., Paull, K., Vistica, D., Hose, C., Langley, J., Cronise, P., Vaigro-Wolff, A., et al. (1991). Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 83, 757–766. - PubMed
- Niculescu-Duvaz, I., Roman, E., Whittaker, S.R., Friedlos, F., Kirk, R., Scanlon, I.J., Davies, L.C., Niculescu-Duvaz, D., Marais, R., and Springer, C.J. (2006). Novel inhibitors of B-RAF based on a disubstituted pyrazine scaffold: Generation of a nanomolar lead. J. Med. Chem. 4, 407–416. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- WT_/Wellcome Trust/United Kingdom
- 13083/CRUK_/Cancer Research UK/United Kingdom
- 080333/Z/06/Z/WT_/Wellcome Trust/United Kingdom
- C107/A10433/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous