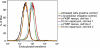Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria - PubMed (original) (raw)
Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria
Margaret S Robinson et al. Dev Cell. 2010.
Abstract
We have developed a method for rapidly inactivating proteins with rapamycin-induced heterodimerization. Cells were stably transfected with siRNA-resistant, FKBP-tagged subunits of the adaptor protein (AP) complexes of clathrin-coated vesicles (CCVs), together with an FKBP and rapamycin-binding domain-containing construct with a mitochondrial targeting signal. Knocking down the endogenous subunit with siRNA, and then adding rapamycin, caused the APs to be rerouted to mitochondria within seconds. Rerouting AP-2 to mitochondria effectively abolished clathrin-mediated endocytosis of transferrin. In cells with rerouted AP-1, endocytosed cation-independent mannose 6-phosphate receptor (CIMPR) accumulated in a peripheral compartment, and isolated CCVs had reduced levels of CIMPR, but normal levels of the lysosomal hydrolase DNase II. Both observations support a role for AP-1 in retrograde trafficking. This type of approach, which we call a "knocksideways," should be widely applicable as a means of inactivating proteins with a time scale of seconds or minutes rather than days.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
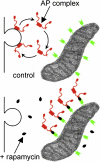
Graphical abstract
Figure 1
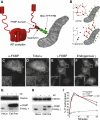
FKBP-Tagged AP Complexes (A and B) Schematic diagrams of the strategy that was used. (C–F) Stable cell lines expressing either α-FKBP (C and D) or γ-FKBP (E and F) were mixed with nontransfected cells and double labeled for the construct and either total α or endogenous γ (using a species-specific antibody). The insets show higher magnification views of the areas inside the boxes. Scale bar, 20 μm. (G and H) Homogenates of control HeLa cells and cell lines expressing either α-FKBP (G) or γ-FKBP (H) were depleted of the endogenous subunit, and western blots were probed with antibodies that recognize both the endogenous protein and the tagged construct. (I) Control and α-FKBP-expressing cells were depleted of endogenous α using siRNA, and endocytosis of prebound 125I-labeled transferrin (Tf) was measured. Data are presented as the means (±SE) from three independent experiments.
Figure 2
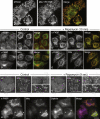
Localization of Constructs and Effect of Rapamycin (A) Cells expressing Mito-YFP-FRB were labeled with an antibody against a mitochondrial marker, MTC02. (B–E) Stable cell lines coexpressing either α-FKBP (B and C) or γ-FKBP (D and E) together with Mito-YFP-FRB were treated with 200 nM rapamycin for either 0 (B and D) or 10 (C and E) min. Electron micrographs of the same conditions are shown in Figure S1. (F and G) Live-cell imaging of cells coexpressing α-FKBP and Mito-YFP-FRB, and transiently transfected with mCherry-tagged σ2. See Movie S1 for the video link. Rapamycin was added to the cells at time 0 (F), which corresponds to 1 min in the video; the frames in (G) show the same cells 9 s later. The panels on the right of (F) and (G) are higher magnification views of the areas inside the boxes. (H) Localization of clathrin in cells with rerouted AP-2. Mixed populations of cells, expressing either α-FKBP only, or coexpressing α-FKBP and Mito-YFP-FRB, were depleted of endogenous α, treated with 200 nM rapamycin for 10 min, and triple labeled for clathrin. Other triple-labeled images are shown in Figure S2. Scale bars, 20 μm.
Figure 3
Assay for AP-2 Function Two different cell lines that stably express both α-FKBP and Mito-YFP-FRB were depleted of endogenous α and allowed to endocytose fluorescently labeled transferrin for 10 min, with or without first treating with rapamycin for 10 min. As a comparison, control cells, either left untreated or depleted of endogenous α, were also allowed to endocytose fluorescent transferrin for 10 min. After stripping off surface-bound transferrin, internalized transferrin was quantified by flow cytometry.
Figure 4
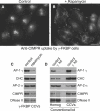
Assays for AP-1 Function (A and B) Cells coexpressing γ-FKBP and Mito-YFP-FRB were depleted of endogenous γ with siRNA, and incubated with anti-CIMPR for 45 min at 37°C, either without (A) or with (B) rapamycin. See also Figure S3. Scale bar, 20 μm. (C) Cells coexpressing γ-FKBP and Mito-YFP-FRB were depleted of endogenous γ with siRNA. Half of the cells were treated with rapamycin for 10 min before homogenization, then CCVs were isolated and western blots probed with the indicated antibodies. (D) Blots of whole-cell homogenates and CCV preparations from a conventional AP-1 γ knockdown, probed with the same antibodies as in (C).
Similar articles
- The aftiphilin/p200/gamma-synergin complex.
Hirst J, Borner GH, Harbour M, Robinson MS. Hirst J, et al. Mol Biol Cell. 2005 May;16(5):2554-65. doi: 10.1091/mbc.e04-12-1077. Epub 2005 Mar 9. Mol Biol Cell. 2005. PMID: 15758025 Free PMC article. - Sorting of major cargo glycoproteins into clathrin-coated vesicles.
Harasaki K, Lubben NB, Harbour M, Taylor MJ, Robinson MS. Harasaki K, et al. Traffic. 2005 Nov;6(11):1014-26. doi: 10.1111/j.1600-0854.2005.00341.x. Traffic. 2005. PMID: 16190982 - Activation of the epidermal growth factor (EGF) receptor induces formation of EGF receptor- and Grb2-containing clathrin-coated pits.
Johannessen LE, Pedersen NM, Pedersen KW, Madshus IH, Stang E. Johannessen LE, et al. Mol Cell Biol. 2006 Jan;26(2):389-401. doi: 10.1128/MCB.26.2.389-401.2006. Mol Cell Biol. 2006. PMID: 16382132 Free PMC article. - The long life of an endocytic patch that misses AP-2.
de León N, Valdivieso MH. de León N, et al. Curr Genet. 2016 Nov;62(4):765-770. doi: 10.1007/s00294-016-0605-3. Epub 2016 Apr 28. Curr Genet. 2016. PMID: 27126383 Review. - Cargo recognition during clathrin-mediated endocytosis: a team effort.
Sorkin A. Sorkin A. Curr Opin Cell Biol. 2004 Aug;16(4):392-9. doi: 10.1016/j.ceb.2004.06.001. Curr Opin Cell Biol. 2004. PMID: 15261671 Review.
Cited by
- Biomolecular condensation orchestrates clathrin-mediated endocytosis in plants.
Dragwidge JM, Wang Y, Brocard L, De Meyer A, Hudeček R, Eeckhout D, Grones P, Buridan M, Chambaud C, Pejchar P, Potocký M, Winkler J, Vandorpe M, Serre N, Fendrych M, Bernard A, De Jaeger G, Pleskot R, Fang X, Van Damme D. Dragwidge JM, et al. Nat Cell Biol. 2024 Mar;26(3):438-449. doi: 10.1038/s41556-024-01354-6. Epub 2024 Feb 12. Nat Cell Biol. 2024. PMID: 38347182 Free PMC article. - The fifth adaptor protein complex.
Hirst J, Barlow LD, Francisco GC, Sahlender DA, Seaman MN, Dacks JB, Robinson MS. Hirst J, et al. PLoS Biol. 2011 Oct;9(10):e1001170. doi: 10.1371/journal.pbio.1001170. Epub 2011 Oct 11. PLoS Biol. 2011. PMID: 22022230 Free PMC article. - Cell-permeant and photocleavable chemical inducer of dimerization.
Zimmermann M, Cal R, Janett E, Hoffmann V, Bochet CG, Constable E, Beaufils F, Wymann MP. Zimmermann M, et al. Angew Chem Int Ed Engl. 2014 Apr 25;53(18):4717-20. doi: 10.1002/anie.201310969. Epub 2014 Mar 26. Angew Chem Int Ed Engl. 2014. PMID: 24677313 Free PMC article. - Role of AP1 and Gadkin in the traffic of secretory endo-lysosomes.
Laulagnier K, Schieber NL, Maritzen T, Haucke V, Parton RG, Gruenberg J. Laulagnier K, et al. Mol Biol Cell. 2011 Jun 15;22(12):2068-82. doi: 10.1091/mbc.E11-03-0193. Epub 2011 Apr 27. Mol Biol Cell. 2011. PMID: 21525240 Free PMC article. - How to Train a Cell-Cutting-Edge Molecular Tools.
Czapiński J, Kiełbus M, Kałafut J, Kos M, Stepulak A, Rivero-Müller A. Czapiński J, et al. Front Chem. 2017 Mar 10;5:12. doi: 10.3389/fchem.2017.00012. eCollection 2017. Front Chem. 2017. PMID: 28344971 Free PMC article. Review.
References
- Ball C.L., Hunt S.P., Robinson M.S. Expression and localisation of α-adaptin isoforms. J. Cell Sci. 1995;108:2865–2875. - PubMed
- Bayle J.H., Grimley J.S., Stankunas K., Gestwicki J.E., Wandless T.J., Crabtree G.R. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem. Biol. 2006;13:99–107. - PubMed
- Bear J.E., Loureiro J.J., Libova I., Fässler R., Wehland J., Gertler F.B. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous