MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele - PubMed (original) (raw)
MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele
Austin T Thiel et al. Cancer Cell. 2010.
Abstract
Oncogenic fusion proteins are capable of initiating tumorigenesis, but the role of their wild-type counterparts in this process is poorly understood. The mixed lineage leukemia (MLL) gene undergoes chromosomal translocations, resulting in the formation of oncogenic MLL fusion proteins (MLL-FPs). Here, we show that menin recruits both wild-type MLL and oncogenic MLL-AF9 fusion protein to the loci of HOX genes to activate their transcription. Wild-type MLL not only catalyzes histone methylation at key target genes but also controls distinct MLL-AF9-induced histone methylation. Notably, the wild-type Mll allele is required for MLL-AF9-induced leukemogenesis and maintenance of MLL-AF9-transformed cells. These findings suggest an essential cooperation between an oncogene and its wild-type counterpart in MLL-AF9-induced leukemogenesis.
2010 Elsevier Inc. All rights reserved.
Figures
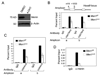
Figure 1. Menin is required for both H3K4 trimethylation and H3K79 dimethylation at Hoxa9 in MA9-transformed cells
(A) Western blot for menin in control or Men1 excised MA9-transformed AT1 cells, which harbored Men1f/f;Cre-ER. The cells were treated with either control DMSO (Men1f/f) or 4-OHT (_Men1_Δ/Δ) to excise the floxed Men1. (B–D) ChIP assay, with two distinct amplicons, for detecting dimethylated H3K79 (B), trimethylated H3K4 (C), and menin binding (D) at Hoxa9 in Men1f/f and _Men1_Δ/Δ AT1 cells. Error bars denote +/− SD.
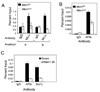
Figure 2. Wt MLL and MA9 are recruited to Hoxa9 in a menin-dependent manner
AT1 cells were treated with either DMSO (Men1f/f) or 4-OHT (_Men1_Δ/Δ) and processed for ChIP assay with either (A) anti-MLL-C or (B) anti-AF9 C-terminus antibodies. (C) THP-1 cells were transduced with either control scrambled or Men1 shRNA-expressing lentiviruses, and used for ChIP assay with anti-AF9 or anti-Dot1L antibodies. Error bars denote +/− SD. See also Figure S1.
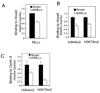
Figure 3. Wt Mll is required for growth of MA9-transformed leukemia cells and expression of Hoxa9 and CCNA2
(A) A diagram for the structure of wt MLL, MA9 fusion protein, and shRNAs targeting various parts of MLL-C but not MA9. AT1 cells were transduced with either vector or MLL-C shRNA 11 retroviruses and monitored for cell number (+/− SD) (B), wt Mll, Hoxa9, and cyclin A (CCNA2) mRNAs (C), and the protein levels of MLL-C and cyclin A (+/− SD) (D). THP-1 cells were transduced with either control scrambled shRNA lentiviruses (Scram) or MLL-C shRNAs. The resulting cells were monitored for change in number (+/− SD) (E), the mRNA levels of wt Mll, Hoxa9, and CCNA2 (+/− SD) (F), and the protein levels of MLL-C and cyclin A (G), MLL-N, MLL-AF9 (MA9), and menin (H). See also Figure S2.
Fig. 4. Wt Mll excision reduces the number of MA9-transformed cells and Hox gene expression
(A) A diagram for the floxed Mll and the primers used to detect the intact or excised Mll. (B) 4-OHT induced excision of the floxed Mll. Spleen cells from a Mllf/f;Cre-ER mouse were cultured with either DMSO or 4-OHT, followed by isolation of the genomic DNA and PCR amplification. (C) A growth curve for MA9-transformed BM cells with either Mllf/f or _Mll_Δ/Δ (+/− SD, cells seeded in triplicate) (D) Quantification of Hoxa9 and Hoxa5 mRNAs in either Mllf/f or _Mll_Δ/Δ MA9-transformed cells (+/− SD). See also Figure S3.
Figure 5. Wt MLL is crucial for maximal methylation of both H3K4 and H3K79 at key target genes
THP-1 cells were transduced with either control scrambled shRNA or MLL-C shRNA 14 lentiviruses to knock down wt MLL and evaluated using ChIP assay for MLL-C binding to Hoxa9 (A) and for histone H3K4m3 and H3K79m2 at Hoxa9 (B) and CCNA2 (C). The specificity of the anti-H3K4m3 antibody and the anti-H3K79m2 were confirmed using specifically modified peptides and Western blot. Error bars denote +/− SD. See also Figure S4.
Figure 6. Wt MLL knockdown suppresses colony formation of MA9-transduced BM
(A) Procedure for the colony formation assay. Bone marrow (BM) cells from a C57B6 mouse were transduced with pMSCV-MA9 retroviruses, and replated in triplicate weekly in methylcellulose medium with G418. After the second plating, surviving MA9 cells were transduced with each of the MLL-C shRNAs (12 and 14) or scrambled vector. (B) A summary of colony numbers for control or Mll shRNA-transduced BM. (C) Representative colonies from the culture plates (Scale bars 5mM). Error bars denote +/− SD.
Figure 7. Wt Mll is required for colony formation of MA9-induced BM
(A) A flowchart for procedures of MA9-induced transformation and 4-OHT-induced Mll excision. (B) Mll excision reduced the number of colonies from MA9-transduced BM from the Mllf/f;Cre-ER mice. (C) 4-OHT failed to reduce colony formation of MA9 retrovirus-transduced BM from the Mllf/f mice. (D) Genotyping with genomic DNA showed that 4-OHT induced Mll excision in MA9-transformed cells with the Mllf/f;Cre-ER genotype (lane 2) but failed to induce Mll excision in MA9-transformed BM cells with Mllf/f but without the Cre-ER transgene (lane 4). (E) Wt Mll excision failed to reduce Hoxa9/Meis1-induced BM colony formation (Top). 4-OHT-induced wt Mll excision in Hoxa9/Meis1-transformed BM (Bottom). (F) Wt Mll excision reduced colony formation from normal BM. BM from corn oil (CO) or TAM-treated Mllf/f;Cre-ER mice was plated on methylcellulose medium and the colony number was scored at the first plating (Top). Wt Mll was excised in BM from TAM-treated, but not from corn oil-fed, Mllf/f;Cre-ER mice (Bottom). (G) BM from corn oil or TAM-treated Mllf/f;Cre-ER mice was first transduced with MA9, followed by plating on methylcellulose medium, and the colony number was scored at the first plating. Error bars denote +/− SD.
Figure 8. Wt Mll is required for MA9-induced leukemogenesis in mice
(A) A diagram for MA9-induced leukemogenesis and wt Mll excision in mice. TAM feeding was done 3 weeks after BM transplantation. (B) The total peripheral blood white blood cells (WBCs) in mice transplanted with MA9-transduced Mllf/f;Cre-ER BM were measured five weeks after corn oil or TAM feeding. (C) Flow cytometry analysis of peripheral blood CD11b+ MA9-transformed donor cells from transplanted mice, five weeks after the mice were fed with corn oil or TAM. (D) Flow cytometry analysis of CD45.2+ splenocytes from either terminally ill and corn oil-fed mice or TAM-fed mice to detect the percentage of c-kit high cells that were FcRγII/III+/CD34+. (E) A summary of the percentage of c-kit high cells that were FcRγII/III+/CD34+ from the corn oil or TAM-fed mice. (F) TAM-induced Mll excision in transplanted MA9-transduced BM in recipient mice increased the survival rate of the mice. Kaplan Meier curve for mice (C57B6/B6-SJL F1) transplanted with MA9-transduced Mllf/f;Cre-ER BM that were fed with either control corn oil (n=11) or TAM (n=7), 3 weeks after MA9 BM transplantation. (G) A model for menin, wt MLL, and MA9/Dot1L tripartite complex-controlled regulation of coupled yet distinct histone methylations, gene transcription, MA9-induced LSCs, and leukemogenesis. See also Figure S5.
Similar articles
- CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis.
Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. Tan J, et al. Cancer Cell. 2011 Nov 15;20(5):563-75. doi: 10.1016/j.ccr.2011.09.008. Cancer Cell. 2011. PMID: 22094252 Free PMC article. - The trithorax protein partner menin acts in tandem with EZH2 to suppress C/EBPα and differentiation in MLL-AF9 leukemia.
Thiel AT, Feng Z, Pant DK, Chodosh LA, Hua X. Thiel AT, et al. Haematologica. 2013 Jun;98(6):918-27. doi: 10.3324/haematol.2012.074195. Epub 2013 Jan 24. Haematologica. 2013. PMID: 23349306 Free PMC article. - DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis.
Nguyen AT, Taranova O, He J, Zhang Y. Nguyen AT, et al. Blood. 2011 Jun 23;117(25):6912-22. doi: 10.1182/blood-2011-02-334359. Epub 2011 Apr 26. Blood. 2011. PMID: 21521783 Free PMC article. - Two decades of leukemia oncoprotein epistasis: the MLL1 paradigm for epigenetic deregulation in leukemia.
Li BE, Ernst P. Li BE, et al. Exp Hematol. 2014 Dec;42(12):995-1012. doi: 10.1016/j.exphem.2014.09.006. Epub 2014 Sep 28. Exp Hematol. 2014. PMID: 25264566 Free PMC article. Review. - Menin as a hub controlling mixed lineage leukemia.
Thiel AT, Huang J, Lei M, Hua X. Thiel AT, et al. Bioessays. 2012 Sep;34(9):771-80. doi: 10.1002/bies.201200007. Epub 2012 Jul 24. Bioessays. 2012. PMID: 22829075 Free PMC article. Review.
Cited by
- Musashi2 sustains the mixed-lineage leukemia-driven stem cell regulatory program.
Park SM, Gönen M, Vu L, Minuesa G, Tivnan P, Barlowe TS, Taggart J, Lu Y, Deering RP, Hacohen N, Figueroa ME, Paietta E, Fernandez HF, Tallman MS, Melnick A, Levine R, Leslie C, Lengner CJ, Kharas MG. Park SM, et al. J Clin Invest. 2015 Mar 2;125(3):1286-98. doi: 10.1172/JCI78440. Epub 2015 Feb 9. J Clin Invest. 2015. PMID: 25664853 Free PMC article. - ECSASB2 mediates MLL degradation during hematopoietic differentiation.
Wang J, Muntean AG, Hess JL. Wang J, et al. Blood. 2012 Feb 2;119(5):1151-61. doi: 10.1182/blood-2011-06-362079. Epub 2011 Dec 15. Blood. 2012. PMID: 22174154 Free PMC article. - Restoring MLL reactivates latent tumor suppression-mediated vulnerability to proteasome inhibitors.
Ge M, Li D, Qiao Z, Sun Y, Kang T, Zhu S, Wang S, Xiao H, Zhao C, Shen S, Xu Z, Liu H. Ge M, et al. Oncogene. 2020 Sep;39(36):5888-5901. doi: 10.1038/s41388-020-01408-7. Epub 2020 Jul 30. Oncogene. 2020. PMID: 32733069 Free PMC article. - Moonlighting with WDR5: A Cellular Multitasker.
Guarnaccia AD, Tansey WP. Guarnaccia AD, et al. J Clin Med. 2018 Jan 30;7(2):21. doi: 10.3390/jcm7020021. J Clin Med. 2018. PMID: 29385767 Free PMC article. Review. - Biological Functions of the KDM2 Family of Histone Demethylases.
Andricovich J, Tzatsos A. Andricovich J, et al. Adv Exp Med Biol. 2023;1433:51-68. doi: 10.1007/978-3-031-38176-8_3. Adv Exp Med Biol. 2023. PMID: 37751135
References
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. - PubMed
- Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316:600–604. - PubMed
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA113962/CA/NCI NIH HHS/United States
- R01 CA113962-05/CA/NCI NIH HHS/United States
- T32 CA009140/CA/NCI NIH HHS/United States
- R01 CA100912/CA/NCI NIH HHS/United States
- T32 CA09140/CA/NCI NIH HHS/United States
- R01 CA100912-05/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous