Organotypic specificity of key RET adaptor-docking sites in the pathogenesis of neurocristopathies and renal malformations in mice - PubMed (original) (raw)
. 2010 Mar;120(3):778-90.
doi: 10.1172/JCI41619. Epub 2010 Feb 15.
Affiliations
- PMID: 20160347
- PMCID: PMC2827965
- DOI: 10.1172/JCI41619
Organotypic specificity of key RET adaptor-docking sites in the pathogenesis of neurocristopathies and renal malformations in mice
Sanjay Jain et al. J Clin Invest. 2010 Mar.
Abstract
The receptor tyrosine kinase ret protooncogene (RET) is implicated in the pathogenesis of several diseases and in several developmental defects, particularly those in neural crest-derived structures and the genitourinary system. In order to further elucidate RET-mediated mechanisms that contribute to these diseases and decipher the basis for specificity in the pleiotropic effects of RET, we characterized development of the enteric and autonomic nervous systems in mice expressing RET9 or RET51 isoforms harboring mutations in tyrosine residues that act as docking sites for the adaptors Plcgamma, Src, Shc, and Grb2. Using this approach, we found that development of the genitourinary system and the enteric and autonomic nervous systems is dependent on distinct RET-stimulated signaling pathways. Thus, mutation of RET51 at Y1062, a docking site for multiple adaptor proteins including Shc, caused distal colon aganglionosis reminiscent of Hirschsprung disease (HSCR). On the other hand, this mutation in RET9, which encodes an isoform that lacks the Grb2 docking site present in RET51, produced severe abnormalities in multiple organs. Mutations that abrogate RET-Plcgamma binding, previously shown to produce features reminiscent of congenital anomalies of kidneys or urinary tract (CAKUT) syndrome, produced only minor abnormalities in the nervous system. Abrogating RET51-Src binding produced no major defects in these systems. These studies provide insight into the basis of organotypic specificity and redundancy in RET signaling within these unique systems and in diseases such as HSCR and CAKUT.
Figures
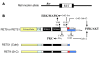
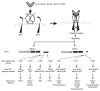
Figure 1. Schematic of the various _RET_-mutant mice used in this study.
(A) Simplified diagram of mutant or WT human RET cDNAs homologously recombined into exon 1 (black box) of the mouse Ret locus using knockout-knockin approach (29). (B) Schematic of the different WT RET9, RET51, and their respective mutant knocked in alleles is shown. The different domains and sizes of RET9 and RET51 are indicated. The area of divergence between the 2 RET isoforms is indicated in red or blue. Also indicated are the key docking tyrosine (Y) residues, the major intracellular adapters that dock at these tyrosines, and the downstream signaling cascades. Homozygous mice were generated that harbor Tyr-to-Phe (Y to F) mutations for each of the indicated Tyr except Y1096. In the RET51(Cdel) allele, residues 1063–1072 of RET9 were replaced with residues 1063–1072 of RET51. This results in a receptor that is essentially RET51 with a deletion of residues 1073–1114, including Y1096. In the RET9(51C) allele, residues 1063–1072 of RET51 were replaced with residues 1063–1072 of RET9. This results in a receptor that is essentially similar to RET9 with C terminus of RET51, including Y1096. Note Grb2 can directly bind to RET51 and also indirectly to Y1062 in both RET9 and RET51.
Figure 2. RET51 is sufficient to support ENS and sympathetic nervous system development and is necessary for the development of cranial parasympathetic ganglia.
(A) RET9 and RET51 can both support normal ENS development. Whole-mount AChE staining (to visualize ENS formation) from WT or the indicated RET51 and RET9 mice (at postnatal day zero, P0) show typical dense reticular, honeycomb-like distal colon staining, indicating normal ENS colonization by both RET isoforms. (B) Redundant roles of RET isoforms in development of sympathetic ganglia. Whole-mount TH immunohistochemistry (brown color) shows normally located SCG and normal projections to the eye (white arrowhead) and submandibular gland (SM, black arrowheads). (C) RET9 alone is unable to fully support parasympathetic ganglia development. Nissl-stained sections of head from P0 mice show normal location of SPG in WT and RET51 (arrows) mice, but a subset of RET9 mice show unilateral SPG agenesis (dashed oval) (see E–H for orientation). (D) Reduced harderian gland innervation in RET9 mice (1- to 2-month-old mice) was observed with anti-TuJ1 immunohistochemistry (red fibers). While TuJ1-positive fibers surround almost all acini in WT and RET51 mice, RET9 harderian glands show reduced innervation consistent with incompletely penetrant unilateral agenesis. (E–H) Anatomical and histological landmarks for SPG analysis. Vertical bar in E shows approximate level of histological sections. (F) Coronal section showing SPG location (white arrow); boxed area is shown at higher magnification in G, which also represents images in C and Figure 7A. (H) Closer view of SPG of one side denotes approximate representation of images in Figure 7B. Scale bars: 400 μm (A); 600 μm (B); 200 μm (C); 100 μm (D); 50 μm (inset); F, 1 mm. br, brain; sep, septum; mu, eye muscles; np, nasopharynx; mn, maxillary nerves; oa, orbital artery; ov, orbital vein; spa, sphenopalatine ganglion.
Figure 3. Essential role of the RET-Shc multidocking site in distal colon innervation.
To determine roles of individual RET adaptor sites in ENS development, AChE whole-mount staining to visualize the ENS was performed on intestines (P0) of the indicated adaptor mutants in the context of RET51 isoform (Y981F, Src adaptor; Y1015F, Plcγ adaptor; Y1062F, Shc adaptor; or null, KO). Neuronal plexus and ganglion formation, depicted by typical reticular, honeycomb like staining pattern at ileocecal (IC) junction and distal colon, occurs in all adaptor mutants except for RET51(Y1062F). The RET51(Y1062F) image shows distal colon with absent enteric ganglia and plexus, but readily visible thick extrinsic nerve fiber bundles similar to those observed in _Ret_-null mice. Varying degrees of colon aganglionosis were seen in RET51(Y1062F) mice. Compared with aganglionosis in terminal ileum (TI) of _Ret_-KO mice, AChE staining of the intestines in RET51(Y1062F) mice detected both a neuronal plexus and ganglia in the TI. The schematic summarizes the extent of bowel colonization by ENS precursors in mutant mice. For RET51(Y1062F), each pentagon represents the location of the most distal enteric ganglion cell in an individual mutant; for other mouse lines, single pentagons represent the entire group, since none of these mice had bowel aganglionosis (refer to graph in Figure 4B for number of mice analyzed for each mutant mouse and associated ENS abnormality). Scale bars: 600 μm (IC junction); 400 μm (distal colon).
Figure 4. Severe ENS defects in RET tyrosine docking mutants lacking the Grb2-binding site.
(A) Whole-mount AChE staining was used to visualize the ENS in P0 intestines in RET9 or the indicated RET9 docking tyrosine mutant mice. Regions of the bowel from the distal colon and duodenum are shown. WT RET9 monoisoformic animals have a normal-appearing dense reticular staining pattern in the duodenum and the distal colon, while RET9(Y981F) (Src mutants) have colon aganglionosis but normal duodenum staining. RET9(Y1015F) (Plcγ) mice have neuronal ganglia and plexus in the colon but at reduced density (hypoganglionosis); duodenum innervation is normal. RET9(Y1062F) mice manifest complete intestinal aganglionosis, as no neurons are present in colon or in duodenum. A compound isoformic Y1062F mutant (RET9/51[Y1062F]) has intermediate aganglionosis compared with RET51(Y1062F) (Figure 3) and RET9(Y1062F) mice, indicating isoform dosage influences intestinal innervation. (B) The bar graph summarizes the innervation phenotypes of all the RET9 and RET51 WT and adaptor mutants examined. Numbers in the bars represent the number of mice with that phenotype. Scale bars: 600 μm (duodenum); 400 μm (distal colon).
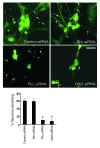
Figure 5. Grb2 and Plcγ are essential for maintaining normal enteric neuron number.
Roles of Grb2, Plcγ, and Shc adaptor proteins in regulating enteric neuron number were examined in vitro. ENS precursors were harvested from E14.5 rat intestines using p75NTR immunoselection and propagated in culture for 7 days. Indicated siRNAs or control (scrambled siRNA) were delivered by lentivirus infection 1 day after plating the cells. Tau1 immunofluorescence was used to detect neurons and their neurites (arrows). Shc siRNA did not affect cell number compared with control. _Plc_γ and Grb2 siRNA caused a severe reduction in neuron numbers. The y axis represents neurons remaining relative to the beginning of the culture. The graph at the bottom shows quantification results from 3 independent experiments (mean ± SD, *P < 0.05 versus control). Scale bar: 25 μm.
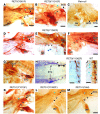
Figure 6. Sympathetic nervous system defects in RET mutants lacking Shc and Grb2 binding sites.
Whole-mount TH immunohistochemistry was performed on P0 pups to assess sympathetic nervous system development in RET9 and RET51 signaling mutants. (A–C) Normal sympathetic nervous system develops in (A) RET9(Y981F) (Src mutant) and (B) RET9(Y1015F) (Plcγ mutant) mice, as demonstrated by normal SCG location and normal innervation to submandibular gland (SM, black arrowheads) and the eye (blue arrowhead). (C) _Ret_-null mice show abnormal caudal location of the SCG, next to the stellate ganglion (STG) instead of the expected normal location (dashed oval); innervations to the SM or the eye are absent. (D–H) RET9(Y1062F) adaptor mutants (lack Shc- and Grb2-binding sites) show a spectrum of sympathetic nervous system defects albeit milder than _Ret_-null animals. These include normal SCG location and projections to eye (blue arrowhead) and submandibular gland (black arrowhead in D), failure of SCG to migrate normally (E and F; dashed circle depicts expected location), mislocalization of the SCG near the stellate ganglion (STG) in G, and fusion of rostral sympathetic chain ganglia (black arrowhead) with the STG, resulting in a gap in the sympathetic chain (double arrowhead) in H. (I) Sympathetic chain (sc) ganglia in RET9(Y1062F) mice are small (black arrows), or absent (dashed oval) and have diminished to absent axonal outgrowths compared with WT mice (blue arrows) in J. (K–M) Docking site mutations in the context of RET51 do not disrupt sympathetic nervous system development. Representative pictures are shown for the indicated adaptor mutants in RET51 context, highlighting normal sympathetic ganglia development. Scale bars: 600 μm (A–H and K–M); 400 μm (I, J).
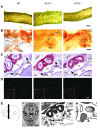
Figure 7. Severe parasympathetic nervous system defects in RET9 docking tyrosine mutant mice.
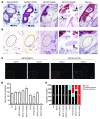
Cranial parasympathetic SPG and their innervation of the harderian gland using Nissl staining (P0 pups) and TuJ1 immunohistochemistry (1- to 2-month-old mice), respectively. Refer to Figure 2, E–H, for anatomical landmarks for SPG and key for annotations. (A and B) RET9(Y1062F) mice exhibit (n = 14) bilateral SPG agenesis, a phenotype similar to that of _Ret_-null mice (dashed oval, expected normal SPG site); RET51(Y1062F) SPG were normally located (arrows in A and B). RET9(Y981F) and RET9(Y1015F) mice showed incomplete penetrance of SPG agenesis, including normal SPG location bilaterally (arrows in A), unilateral, or bilateral SPG agenesis (SPG agenesis indicated by dashed ovals in B and summarized in E). (C) Differences in innervations (TuJ1 staining, red fibers surrounding each acinus) of the 2 harderian glands from the same mutant mouse. Note that gland 1 of each mutant had TuJ1-positive fibers surrounding all acini, but gland 2 innervation was markedly decreased or absent, consistent with SPG unilateral agenesis (closer view shown in the inset). (D) Summary of SPG neuronal numbers in RET mutants, in which SPG are formed. Quantification of neuron numbers in SPG show no significant differences in mutant and WT mice except RET9(Y1062F) mice, which had SPG agenesis. The graph depicts number of neurons in each completely sectioned SPG (mean ± SEM; numbers at the bottom denote SPG used per genotype). (E) Spectrum of SPG abnormalities observed in _RET_-mutant mice. Results of SPG location from both RET51 and RET9 mutant mice are summarized in the bar graph. Numbers in each colored bar represent the number of SPG with the corresponding defect. Scale bars: 200 μm (A); 50 μm (B); 100 μm (C); 50 μm (inset).
Figure 8. Summary model depicting the main tissues affected by key docking tyrosines activated by GFL-RET signaling.
One of the 4 GFLs binds 1 of the 4 GFRα coreceptors and forms a multimeric complex with RET leading to receptor activation. The phosphorylated docking sites on each of the major isoforms, RET9 and RET51, interact with the indicated adaptors and the signaling cascades. RET9 lacks the extra GRB2-binding site (Y1096), and defects in general are more severe and affect more systems when the docking sites are mutated in RET9 context. Asterisks on the MAPK/PI3K pathways denote partial reduction due to redundant activation through intact Y1062 or Y1096.
Similar articles
- Critical and distinct roles for key RET tyrosine docking sites in renal development.
Jain S, Encinas M, Johnson EM Jr, Milbrandt J. Jain S, et al. Genes Dev. 2006 Feb 1;20(3):321-33. doi: 10.1101/gad.1387206. Genes Dev. 2006. PMID: 16452504 Free PMC article. - Distinct turnover of alternatively spliced isoforms of the RET kinase receptor mediated by differential recruitment of the Cbl ubiquitin ligase.
Scott RP, Eketjäll S, Aineskog H, Ibáñez CF. Scott RP, et al. J Biol Chem. 2005 Apr 8;280(14):13442-9. doi: 10.1074/jbc.M500507200. Epub 2005 Jan 27. J Biol Chem. 2005. PMID: 15677445 - Two distinct mutations of the RET receptor causing Hirschsprung's disease impair the binding of signalling effectors to a multifunctional docking site.
Geneste O, Bidaud C, De Vita G, Hofstra RM, Tartare-Deckert S, Buys CH, Lenoir GM, Santoro M, Billaud M. Geneste O, et al. Hum Mol Genet. 1999 Oct;8(11):1989-99. doi: 10.1093/hmg/8.11.1989. Hum Mol Genet. 1999. PMID: 10484767 - Developmental determinants of the independence and complexity of the enteric nervous system.
Gershon MD. Gershon MD. Trends Neurosci. 2010 Oct;33(10):446-56. doi: 10.1016/j.tins.2010.06.002. Epub 2010 Jul 13. Trends Neurosci. 2010. PMID: 20633936 Review. - Hirschsprung's disease as a model of complex genetic etiology.
Borrego S, Ruiz-Ferrer M, Fernández RM, Antiñolo G. Borrego S, et al. Histol Histopathol. 2013 Sep;28(9):1117-36. doi: 10.14670/HH-28.1117. Epub 2013 Apr 19. Histol Histopathol. 2013. PMID: 23605783 Review.
Cited by
- The developmental etiology and pathogenesis of Hirschsprung disease.
Butler Tjaden NE, Trainor PA. Butler Tjaden NE, et al. Transl Res. 2013 Jul;162(1):1-15. doi: 10.1016/j.trsl.2013.03.001. Epub 2013 Mar 22. Transl Res. 2013. PMID: 23528997 Free PMC article. Review. - Multidisciplinary approaches for elucidating genetics and molecular pathogenesis of urinary tract malformations.
Khan K, Ahram DF, Liu YP, Westland R, Sampogna RV, Katsanis N, Davis EE, Sanna-Cherchi S. Khan K, et al. Kidney Int. 2022 Mar;101(3):473-484. doi: 10.1016/j.kint.2021.09.034. Epub 2021 Nov 12. Kidney Int. 2022. PMID: 34780871 Free PMC article. Review. - MiR-195-5p inhibits proliferation and invasion of nerve cells in Hirschsprung disease by targeting GFRA4.
Wang G, Wang H, Zhang L, Guo F, Wu X, Liu Y. Wang G, et al. Mol Cell Biochem. 2021 May;476(5):2061-2073. doi: 10.1007/s11010-021-04055-y. Epub 2021 Jan 30. Mol Cell Biochem. 2021. PMID: 33515383 - Retrograde Ret signaling controls sensory pioneer axon outgrowth.
Tuttle A, Drerup CM, Marra M, McGraw H, Nechiporuk AV. Tuttle A, et al. Elife. 2019 Sep 2;8:e46092. doi: 10.7554/eLife.46092. Elife. 2019. PMID: 31476133 Free PMC article. - Regulation of Renal Differentiation by Trophic Factors.
Kurtzeborn K, Cebrian C, Kuure S. Kurtzeborn K, et al. Front Physiol. 2018 Nov 12;9:1588. doi: 10.3389/fphys.2018.01588. eCollection 2018. Front Physiol. 2018. PMID: 30483151 Free PMC article. Review.
References
- Ponder BA, Smith D. The MEN II syndromes and the role of the ret proto-oncogene. Adv Cancer Res. 1996;70:179–222. - PubMed
- Amiel J, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45(1):1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DK6459201/DK/NIDDK NIH HHS/United States
- R01 DK082531/DK/NIDDK NIH HHS/United States
- DK57038/DK/NIDDK NIH HHS/United States
- P30-DK079333/DK/NIDDK NIH HHS/United States
- K08 HD047396-04/HD/NICHD NIH HHS/United States
- R01 DK081644/DK/NIDDK NIH HHS/United States
- R01 DK057038/DK/NIDDK NIH HHS/United States
- R01 DK082531-01/DK/NIDDK NIH HHS/United States
- P30 DK079333/DK/NIDDK NIH HHS/United States
- R01 DK081644-01A1/DK/NIDDK NIH HHS/United States
- DK081644/DK/NIDDK NIH HHS/United States
- K08 HD047396-05/HD/NICHD NIH HHS/United States
- AG013730/AG/NIA NIH HHS/United States
- R01 AG013730/AG/NIA NIH HHS/United States
- DK082531/DK/NIDDK NIH HHS/United States
- K08 HD047396/HD/NICHD NIH HHS/United States
- K08 HD047396-03/HD/NICHD NIH HHS/United States
- HD047396/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous