PANGEA: pipeline for analysis of next generation amplicons - PubMed (original) (raw)
doi: 10.1038/ismej.2010.16. Epub 2010 Feb 25.
David B Crabb, Austin G Davis-Richardson, Diane Chauliac, Jennifer M Mobberley, Kelsey A Gano, Nabanita Mukherjee, George Casella, Luiz F W Roesch, Brandon Walts, Alberto Riva, Gary King, Eric W Triplett
Affiliations
- PMID: 20182525
- PMCID: PMC2974434
- DOI: 10.1038/ismej.2010.16
PANGEA: pipeline for analysis of next generation amplicons
Adriana Giongo et al. ISME J. 2010 Jul.
Abstract
High-throughput DNA sequencing can identify organisms and describe population structures in many environmental and clinical samples. Current technologies generate millions of reads in a single run, requiring extensive computational strategies to organize, analyze and interpret those sequences. A series of bioinformatics tools for high-throughput sequencing analysis, including pre-processing, clustering, database matching and classification, have been compiled into a pipeline called PANGEA. The PANGEA pipeline was written in Perl and can be run on Mac OSX, Windows or Linux. With PANGEA, sequences obtained directly from the sequencer can be processed quickly to provide the files needed for sequence identification by BLAST and for comparison of microbial communities. Two different sets of bacterial 16S rRNA sequences were used to show the efficiency of this workflow. The first set of 16S rRNA sequences is derived from various soils from Hawaii Volcanoes National Park. The second set is derived from stool samples collected from diabetes-resistant and diabetes-prone rats. The workflow described here allows the investigator to quickly assess libraries of sequences on personal computers with customized databases. PANGEA is provided for users as individual scripts for each step in the process or as a single script where all processes, except the chi(2) step, are joined into one program called the 'backbone'.
Figures
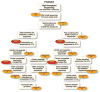
Figure 1
PANGEA workflow. Overview of the pipeline analyses that flow from the raw datasets to the χ2 and Shannon diversity index results.
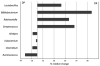
Figure 2
Percent relative change of eight bacterial genera that differ statistically in the χ2-test (_P_-value≤0.01) in the BB-DP and BB-DR rat stool samples.
Similar articles
- A comprehensive evaluation of the sl1p pipeline for 16S rRNA gene sequencing analysis.
Whelan FJ, Surette MG. Whelan FJ, et al. Microbiome. 2017 Aug 14;5(1):100. doi: 10.1186/s40168-017-0314-2. Microbiome. 2017. PMID: 28807046 Free PMC article. - Using QIIME to analyze 16S rRNA gene sequences from microbial communities.
Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Kuczynski J, et al. Curr Protoc Microbiol. 2012 Nov;Chapter 1:Unit 1E.5.. doi: 10.1002/9780471729259.mc01e05s27. Curr Protoc Microbiol. 2012. PMID: 23184592 Free PMC article. - Diazotroph Community Characterization via a High-Throughput nifH Amplicon Sequencing and Analysis Pipeline.
Gaby JC, Rishishwar L, Valderrama-Aguirre LC, Green SJ, Valderrama-Aguirre A, Jordan IK, Kostka JE. Gaby JC, et al. Appl Environ Microbiol. 2018 Jan 31;84(4):e01512-17. doi: 10.1128/AEM.01512-17. Print 2018 Feb 15. Appl Environ Microbiol. 2018. PMID: 29180374 Free PMC article. - Generating amplicon reads for microbial community assessment with next-generation sequencing.
Gołębiewski M, Tretyn A. Gołębiewski M, et al. J Appl Microbiol. 2020 Feb;128(2):330-354. doi: 10.1111/jam.14380. Epub 2019 Aug 1. J Appl Microbiol. 2020. PMID: 31299126 Review. - High-throughput identification and quantification of bacterial cells in the microbiota based on 16S rRNA sequencing with single-base accuracy using BarBIQ.
Jin J, Yamamoto R, Shiroguchi K. Jin J, et al. Nat Protoc. 2024 Jan;19(1):207-239. doi: 10.1038/s41596-023-00906-8. Epub 2023 Nov 27. Nat Protoc. 2024. PMID: 38012397 Review.
Cited by
- An analysis of thaumarchaeota populations from the northern gulf of Mexico.
Tolar BB, King GM, Hollibaugh JT. Tolar BB, et al. Front Microbiol. 2013 Apr 9;4:72. doi: 10.3389/fmicb.2013.00072. eCollection 2013. Front Microbiol. 2013. PMID: 23577005 Free PMC article. - Characterization of the Relative Abundance of the Citrus Pathogen Ca. Liberibacter Asiaticus in the Microbiome of Its Insect Vector, Diaphorina citri, using High Throughput 16S rRNA Sequencing.
Fagen JR, Giongo A, Brown CT, Davis-Richardson AG, Gano KA, Triplett EW. Fagen JR, et al. Open Microbiol J. 2012;6:29-33. doi: 10.2174/1874285801206010029. Epub 2012 Apr 6. Open Microbiol J. 2012. PMID: 22529882 Free PMC article. - Assessment of soil bacterial communities in integrated crop production systems within the Amazon Biome, Brazil: a comparative study.
do Carmo KB, Dias R, de Quadros PD, Berber GCM, Bourscheidt MLB, de Farias Neto AL, Dos Santos Weber OL, Triplett EW, Ferreira A. do Carmo KB, et al. Braz J Microbiol. 2024 Sep;55(3):2815-2825. doi: 10.1007/s42770-024-01352-8. Epub 2024 May 2. Braz J Microbiol. 2024. PMID: 38696039 Free PMC article. - Patterns and processes of microbial community assembly.
Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S. Nemergut DR, et al. Microbiol Mol Biol Rev. 2013 Sep;77(3):342-56. doi: 10.1128/MMBR.00051-12. Microbiol Mol Biol Rev. 2013. PMID: 24006468 Free PMC article. Review. - CANGS DB: a stand-alone web-based database tool for processing, managing and analyzing 454 data in biodiversity studies.
Pandey RV, Nolte V, Boenigk J, Schlötterer C. Pandey RV, et al. BMC Res Notes. 2011 Jun 30;4:227. doi: 10.1186/1756-0500-4-227. BMC Res Notes. 2011. PMID: 21718534 Free PMC article.
References
- Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, et al. Microbial community structure in the North Pacific Ocean. ISME J. 2009;3:1374–1386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials