Combinatorial profiling of chromatin binding modules reveals multisite discrimination - PubMed (original) (raw)
Combinatorial profiling of chromatin binding modules reveals multisite discrimination
Adam L Garske et al. Nat Chem Biol. 2010 Apr.
Abstract
Specific interactions between post-translational modifications (PTMs) and chromatin-binding proteins are central to the idea of a 'histone code'. Here, we used a 5,000-member, PTM-randomized, combinatorial peptide library based on the N terminus of histone H3 to interrogate the multisite specificity of six chromatin binding modules, which read the methylation status of Lys4. We found that Thr3 phosphorylation, Arg2 methylation and Thr6 phosphorylation are critical additional PTMs that modulate the ability to recognize and bind histone H3. Notably, phosphorylation of Thr6 yielded the most varied effect on protein binding, suggesting an important regulatory mechanism for readers of the H3 tail. Mass spectrometry and antibody-based evidence indicate that this previously uncharacterized modification exists on native H3, and NMR analysis of ING2 revealed the structural basis for discrimination. These investigations reveal a continuum of binding affinities in which multisite PTM recognition involves both switch- and rheostat-like properties, yielding graded effects that depend on the inherent 'reader' specificity.
Conflict of interest statement
Competing financial interests
J.M.D and A.L.G have a patent pending (No. 11/585,625) that describes the construction and uses of PTM peptide libraries.
Figures
Figure 1. PTMs included in the 5000- member combinatorial H3 tail peptide library
(a) Positions of randomization are annotated by ‘X’. Arginines 2 and 8 can be unmodified, monomethylated, symmetrically or asymmetrically dimethylated or citrullinated. Lysines 4 and 9 can be unmodified, mono-, di-, or trimethylated or acetylated. Threonines 3 and 6 as well as serine 10 can be unmodified or phosphorylated. Peptides are tethered to a solid-support via a linker comprised of methionine, arginine and a PEG spacer. (b) A digital image of the library screen for the JMJD2A double tudor domain. Dark blue beads are marked with a circle (○) and colorless beads with a rectangle (□).
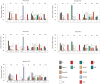
Figure 2. Graphical depiction of discrimination factors obtained from H3 library screens
Values for discrimination factors were obtained by dividing the percent frequency of each modification observed in the intensely blue pool for a given screen by the percent frequency of each corresponding modification from a random group of 100 library members. Discrimination factors represent the fold-likelihood of observing a particular modification in a protein screening experiment relative to random chance. Chi-squared values for each residue are reported along the x-axis. Serine and threonine residues allow for 1 degree of freedom (DF) while lysine and arginine allow for 4 DF. Values above the 99% confidence level for statistical significance are marked with an asterisk (*) (for 1 degree of freedom (DF) = 6.63 and 4 DF = 13.28). A double asterisk (**) is used to denote positions with notably high chi-squared values (above 99.9% confidence level for statistical significance where 1 DF = 10.83 and 4 DF = 18.47).
Figure 3. Detection of H3T6ph by Western blot analysis
(a) Spot blot Western with six distinct H3 peptides residues 1-11 (1:H3T3ph; 2:H3unmod; 3:H3T11ph; 4:H3T6ph; 5:H3K4me3T6ph; 6:H3S10ph). (b) H3T6ph-antibody recognizes H3T6ph from native histones extracted from HeLa cell nuclei (left panel). (c) A 1 μM H3T6ph (1-11) peptide competition diminishes the signal to background levels. The black arrow indicates the band corresponding to H3. The asterisk (*) most likely pertains to H3 C-terminal degradation products.
Figure 4. Detection of H3T6ph using mass spectrometry
(a) Extracted ion chromatogram of the [M+2H]2+ ion at 455.724 m/z, the expected mass of both H3T3phos and H3T6phos from HeLa cells. This sample was subjected to propionic anhydride derivatization, methyl esterification and immobilized metal affinity chromatography (IMAC) for facilitated analysis and phosphopeptide enrichment. As can be see, a major peak is observed at 16.1 minutes, while a second minor resolved peak is clearly visible at 15.8 minutes. Mass accuracy was found to be ~2ppm on either peak as recorded on an Orbitrap mass spectrometer. (b) MS/MS spectrum of the species eluting at 16.1 minutes. The MS/MS fragments show that the sequence is from the 3-8 residues of histone H3 containing T3 phosphorylation. (c) MS/MS spectrum of the species eluting at 15.8 minutes. The MS/MS fragments show that the sequence is from the 3–8 residues of histone H3 containing T6 phosphorylation. Note pr = propionyl amide (56 Da), phos = phosphorylation (80 Da), and —OMe = methyl ester (14 Da).
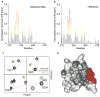
Figure 5. Identification of the H3K4me3T6ph-binding site of the ING2 PHD finger
The histograms show normalized 1H,15N chemical shift changes in backbone amides of the ING2 PHD finger induced by the H3K4me3T6ph (a) and H3K4me3 (b) peptides. The protein:peptide ratio is 1:5, which represents saturation for both interactions. (c) Superimposed 1H,15N heteronuclear single quantum coherence (HSQC) spectra of the ligand-free (black), H3K4me3T6ph-bound (purple) and H3K4me3-bound (green) ING2 PHD finger (0.2 mM). (d) Residues that show a unique pattern of chemical shift perturbations in (c) are colored in red on the surface of the ING2 PHD-H3K4me3 complex and labeled. The residues that exhibit large but parallel chemical shift changes upon addition of either H3K4me3T6ph or H3K4me3 are colored in gray.
Similar articles
- Systems Level Analysis of Histone H3 Post-translational Modifications (PTMs) Reveals Features of PTM Crosstalk in Chromatin Regulation.
Schwämmle V, Sidoli S, Ruminowicz C, Wu X, Lee CF, Helin K, Jensen ON. Schwämmle V, et al. Mol Cell Proteomics. 2016 Aug;15(8):2715-29. doi: 10.1074/mcp.M115.054460. Epub 2016 Jun 14. Mol Cell Proteomics. 2016. PMID: 27302890 Free PMC article. - Application of modified histone peptide arrays in chromatin research.
Mauser R, Jeltsch A. Mauser R, et al. Arch Biochem Biophys. 2019 Jan;661:31-38. doi: 10.1016/j.abb.2018.10.019. Epub 2018 Nov 2. Arch Biochem Biophys. 2019. PMID: 30391375 Review. - Combinations of histone post-translational modifications.
Taylor BC, Young NL. Taylor BC, et al. Biochem J. 2021 Feb 12;478(3):511-532. doi: 10.1042/BCJ20200170. Biochem J. 2021. PMID: 33567070 Review. - Deciphering and engineering chromodomain-methyllysine peptide recognition.
Hard R, Li N, He W, Ross B, Mo GCH, Peng Q, Stein RSL, Komives E, Wang Y, Zhang J, Wang W. Hard R, et al. Sci Adv. 2018 Nov 7;4(11):eaau1447. doi: 10.1126/sciadv.aau1447. eCollection 2018 Nov. Sci Adv. 2018. PMID: 30417094 Free PMC article. - Engineered Reader Proteins for Enhanced Detection of Methylated Lysine on Histones.
Albanese KI, Krone MW, Petell CJ, Parker MM, Strahl BD, Brustad EM, Waters ML. Albanese KI, et al. ACS Chem Biol. 2020 Jan 17;15(1):103-111. doi: 10.1021/acschembio.9b00651. Epub 2019 Nov 1. ACS Chem Biol. 2020. PMID: 31634430 Free PMC article.
Cited by
- The spread of chemical biology into chromatin.
Hegazi E, Muir TW. Hegazi E, et al. J Biol Chem. 2024 Sep 12;300(11):107776. doi: 10.1016/j.jbc.2024.107776. Online ahead of print. J Biol Chem. 2024. PMID: 39276931 Free PMC article. Review. - Genome-Wide Identification and Characterization of the Salvia miltiorrhiza Histone Deacetylase (HDAC) Family in Response to Multiple Abiotic Stresses.
Chen J, Ying Y, Yao L, Xu Z, Yu Z, Kai G. Chen J, et al. Plants (Basel). 2024 Feb 21;13(5):580. doi: 10.3390/plants13050580. Plants (Basel). 2024. PMID: 38475427 Free PMC article. - Interrogating epigenetic mechanisms with chemically customized chromatin.
Hananya N, Koren S, Muir TW. Hananya N, et al. Nat Rev Genet. 2024 Apr;25(4):255-271. doi: 10.1038/s41576-023-00664-z. Epub 2023 Nov 20. Nat Rev Genet. 2024. PMID: 37985791 Review. - Release of Histone H3K4-reading transcription factors from chromosomes in mitosis is independent of adjacent H3 phosphorylation.
Harris RJ, Heer M, Levasseur MD, Cartwright TN, Weston B, Mitchell JL, Coxhead JM, Gaughan L, Prendergast L, Rico D, Higgins JMG. Harris RJ, et al. Nat Commun. 2023 Nov 9;14(1):7243. doi: 10.1038/s41467-023-43115-3. Nat Commun. 2023. PMID: 37945563 Free PMC article. - Clustered PHD domains in KMT2/MLL proteins are attracted by H3K4me3 and H3 acetylation-rich active promoters and enhancers.
Stroynowska-Czerwinska AM, Klimczak M, Pastor M, Kazrani AA, Misztal K, Bochtler M. Stroynowska-Czerwinska AM, et al. Cell Mol Life Sci. 2023 Jan 4;80(1):23. doi: 10.1007/s00018-022-04651-1. Cell Mol Life Sci. 2023. PMID: 36598580 Free PMC article.
References
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. - PubMed
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32GM008505/GM/NIGMS NIH HHS/United States
- GM059785/GM/NIGMS NIH HHS/United States
- R01 GM059785-11/GM/NIGMS NIH HHS/United States
- R37 GM059785/GM/NIGMS NIH HHS/United States
- R01 GM059785/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous