Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence - PubMed (original) (raw)
Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence
Agustin Chicas et al. Cancer Cell. 2010.
Abstract
The RB protein family (RB, p107, and p130) has overlapping and compensatory functions in cell-cycle control. However, cancer-associated mutations are almost exclusively found in RB, implying that RB has a nonredundant role in tumor suppression. We demonstrate that RB preferentially associates with E2F target genes involved in DNA replication and is uniquely required to repress these genes during senescence but not other growth states. Consequently, RB loss leads to inappropriate DNA synthesis following a senescence trigger and, together with disruption of a p21-mediated cell-cycle checkpoint, enables extensive proliferation and rampant genomic instability. Our results identify a nonredundant RB effector function that may contribute to tumor suppression and reveal how loss of RB and p53 cooperate to bypass senescence.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
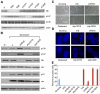
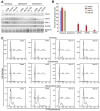
Figure 1. RNAi-mediated knockdown of RB but not p107 or p130 impairs ras-induced senescence
(A) Immunoblots of growing IMR90 cells infected with the indicated shRNAs probed for RB, p107 or p130. Actin was used as loading control. (B) Immunoblots of ras-senescent IMR90 cells. Chromatin-bound fractions were used for the RB, p107 and p130 blots and Histone H3 was used as loading control. Whole cell lysates were used for the p16 and ras blots and actin was used as loading control. (C) SA-β-galactosidase staining. Scale bar=100uM (D) DAPI staining to visualize SAHF. Scale bar=10uM. (E) Quantification of SAHF (red bars), BrdU incorporation (blue bars). Values represent the mean +/− standard error of at least three independent experiments. (V) is empty vector, (S) is senescent, (G) is growing, and (Q) is quiescent.
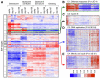
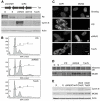
Figure 2. RB represses components of the replication machinery during cellular senescence
(A) Heatmap of expression patterns derived from hierarchical clustering of RB responsive genes (1826 probes) highlighting the E2F target genes. (B-E) Magnification of various gene clusters. (B) Immuno-surveillance cluster (C) Cyclin E cluster (D) DNA replication factors clusters. (E) Mitotic cell cycle cluster. See also Figure S1 and Tables S1-S3.
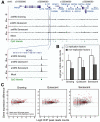
Figure 3. Selective binding of RB to the promoters of DNA replication factors
(A) Binding patterns of RB to the MCM3 gene shown as custom tracks on the UCSC genome browser. (B) Histogram comparing binding of RB-as measured by read counts-to DNA replication factors vs other RB targets. Error bars represent the standard deviation of the read counts. (C) Correlation of RB binding to gene promoters and expression of those genes in RB-deficient cells under different growth conditions. See also Figure S2 and Table S4-S5.
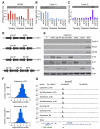
Figure 4. Repression of E2F target genes is unique for RB in senescence but is redundant in quiescence
Immunoblots and qPCR to measure expression of (A) MCM2, (B) Cyclin A, and (C) Cyclin E1 in growing, quiescent or senescent cells expressing the indicated shRNA. qPCR values are averages of representative experiments done in triplicates. (D) Schematic diagram of the polycistronic shRNAs used to knockdown the indicated members of the RB family. (E) Immunoblots probed for the indicated protein from quiescent IMR90 cells lysates infected with the indicated shRNA. (F) Histograms comparing the binding intensity of p130 at gene promoters before (non-shRB) and after RNAi-mediated suppression of RB (shRB) in quiescent (top) and senescent (bottom) cells. The shift to the right of zero indicates that the p130-specific antibody co-immunoprecipitates more promoter DNA in the absence of RB. (G) Binding patterns of p130 and RB to the MCM5 gene shown as custom tracks on the UCSC genome browser. p130 binding to the MCM5 gene promoter increases 1.8 fold in RB-deficient quiescent cells while binding in senescent cells remain unchanged. See also Figure S3 and Table S6
Figure 5. RB is required to prevent inappropriate DNA replication in cells undergoing senescence
(A) Immunoblots of chromatin-bound proteins probed for MCM2, MCM3, ORC1, and Cyclin E1. Blots were normalized by Coomassie Blue staining for histones. (B) Time course BrdU incorporation assays of cells undergoing senescence in the absence of RB, p107 or p130 compared to vector control. (S/PS3, S/PS5, S/PS7 stand for post-selection day 3, 5, or 7 respectively). Values represent the mean +/− standard error of a least three independent experiments. (C) Cells cycle profiles of growing (top), quiescent (middle) or senescent (bottom) IMR90 cells expressing the indicated shRNA. .
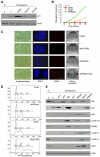
Figure 6. RB repression of Cyclin E1 is required to prevent replication of senescent cells
(A) Immunoblots of lysates from senescent cells expressing a polycistronic shRNA targeting RB and cyclin E1 (Tan depicted on the top of the figure) probed for RB and Cyclin E1. Growing (G), Senescent (S), senescent cells expressing shRNA targeting only RB (shRB/S) or only cyclin E1 (shE1/S) are used as controls. Actin is used as loading control. (B) Cell cycle profiles of vector control senescent cells (V/S), senescent cells expressing shRB (shRB/S) or the tandem shRB/cE1 shRNA (Tan/S). (C) Immunofluorescence of pre-extracted cells to measure proper association of MCM2 with chromatin. Percentage of MCM2 positive cells is indicated in the bottom right corner of the images. DAPI staining was used to visualize the nuclei. At least 200 cells were counted per experiment. Scale bar =100um. (D) Immunoblots of chromatin-bound extracts from growing (G), quiescent (Q), vector control senescent cells (V/S), senescent cells expressing shRB (shRB/S) or the tandem shRB/cE1 shRNA (Tan/S): two independent samples are shown. (E) Immunoblots of lysates from senescent cells expressing the tandem shRB/cE1 shRNA (Tan/S) and shRNA-resistant cyclin E1 cDNA encoding either wild type (WtcE) or kinase-deficient (KDcE) cyclin E1. Lysates from growing (G), vector control senescent cells (V/S), senescent cells expressing shRB (shRB/S) or the tandem shRB/cE1 shRNA (Tan/S) alone are used as controls. See also Figure S4.
Figure 7. RB loss triggers a p53/p21 dependent checkpoint that prevents escape from senescence
(A) Immunoblots of lysates from IMR90 senescent cells expressing the indicated shRNA probed for p21 or actin. (B) Growth curve of ras-infected cells expressing the indicated shRNAs. Counting was initiated at PS7 (post-selection day 7) at which time the vector control cells are fully growth arrested as measure by BrdU incorporation. (C) Analysis of PS7 cells for different proliferation and senescence markers and for the ability to form colonies at low density. Unlike vector control, RB or p21 suppression, suppression of both RB and p21 leads to increase in cell proliferation as measured by BrdU incorporation (20% positive compared to 2% for either shRB or shp21 alone) and colony formation. Scale bars for the micrographs showing SA-β-gal staining and BrdU immunoflourescence=100uM and for DAPI staining=10uM. (D) Cell cycle profiles of ras-infected cells expressing the indicated shRNA. (E) Immunoblots of lysates from ras-infected cells expressing the indicated shRNA probed for the indicated proteins. See also figure S5
Comment in
- What's so special about RB?
Burd CE, Sharpless NE. Burd CE, et al. Cancer Cell. 2010 Apr 13;17(4):313-4. doi: 10.1016/j.ccr.2010.03.010. Cancer Cell. 2010. PMID: 20385355 Free PMC article.
Similar articles
- Distinct roles for p107 and p130 in Rb-independent cellular senescence.
Lehmann BD, Brooks AM, Paine MS, Chappell WH, McCubrey JA, Terrian DM. Lehmann BD, et al. Cell Cycle. 2008 May 1;7(9):1262-8. doi: 10.4161/cc.7.9.5945. Epub 2008 Mar 7. Cell Cycle. 2008. PMID: 18418057 Free PMC article. - RB, p130 and p107 differentially repress G1/S and G2/M genes after p53 activation.
Schade AE, Fischer M, DeCaprio JA. Schade AE, et al. Nucleic Acids Res. 2019 Dec 2;47(21):11197-11208. doi: 10.1093/nar/gkz961. Nucleic Acids Res. 2019. PMID: 31667499 Free PMC article. - Loss of the retinoblastoma tumor suppressor: differential action on transcriptional programs related to cell cycle control and immune function.
Markey MP, Bergseid J, Bosco EE, Stengel K, Xu H, Mayhew CN, Schwemberger SJ, Braden WA, Jiang Y, Babcock GF, Jegga AG, Aronow BJ, Reed MF, Wang JY, Knudsen ES. Markey MP, et al. Oncogene. 2007 Sep 20;26(43):6307-18. doi: 10.1038/sj.onc.1210450. Epub 2007 Apr 23. Oncogene. 2007. PMID: 17452985 - The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis.
Dannenberg JH, te Riele HP. Dannenberg JH, et al. Results Probl Cell Differ. 2006;42:183-225. doi: 10.1007/400_002. Results Probl Cell Differ. 2006. PMID: 16903212 Review. - The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts.
Vélez-Cruz R, Johnson DG. Vélez-Cruz R, et al. Int J Mol Sci. 2017 Aug 16;18(8):1776. doi: 10.3390/ijms18081776. Int J Mol Sci. 2017. PMID: 28812991 Free PMC article. Review.
Cited by
- The requirement for cyclin D function in tumor maintenance.
Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer H, Sicinski P. Choi YJ, et al. Cancer Cell. 2012 Oct 16;22(4):438-51. doi: 10.1016/j.ccr.2012.09.015. Cancer Cell. 2012. PMID: 23079655 Free PMC article. - p63-microRNA feedback in keratinocyte senescence.
Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahé C, Calin GA, Candi E, Melino G. Rivetti di Val Cervo P, et al. Proc Natl Acad Sci U S A. 2012 Jan 24;109(4):1133-8. doi: 10.1073/pnas.1112257109. Epub 2012 Jan 6. Proc Natl Acad Sci U S A. 2012. PMID: 22228303 Free PMC article. - Senescence from G2 arrest, revisited.
Gire V, Dulic V. Gire V, et al. Cell Cycle. 2015;14(3):297-304. doi: 10.1080/15384101.2014.1000134. Cell Cycle. 2015. PMID: 25564883 Free PMC article. Review. - Control of cell cycle transcription during G1 and S phases.
Bertoli C, Skotheim JM, de Bruin RA. Bertoli C, et al. Nat Rev Mol Cell Biol. 2013 Aug;14(8):518-28. doi: 10.1038/nrm3629. Nat Rev Mol Cell Biol. 2013. PMID: 23877564 Free PMC article. Review. - RRM2B suppresses activation of the oxidative stress pathway and is up-regulated by p53 during senescence.
Kuo ML, Sy AJ, Xue L, Chi M, Lee MT, Yen T, Chiang MI, Chang L, Chu P, Yen Y. Kuo ML, et al. Sci Rep. 2012;2:822. doi: 10.1038/srep00822. Epub 2012 Nov 8. Sci Rep. 2012. PMID: 23139867 Free PMC article.
References
- Butt AJ, Caldon CE, McNeil CM, Swarbrick A, Musgrove EA, Sutherland RL. Cell cycle machinery: links with genesis and treatment of breast cancer. Adv Exp Med Biol. 2008;630:189–205. - PubMed
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. - PubMed
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHMI/Howard Hughes Medical Institute/United States
- 5F32AG027631/AG/NIA NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 AG016379-11/AG/NIA NIH HHS/United States
- AG16379/AG/NIA NIH HHS/United States
- R01 AG016379/AG/NIA NIH HHS/United States
- R56 AG016379/AG/NIA NIH HHS/United States
- F32 AG027631/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous