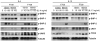In vivo regulation of the allergic response by the IL-4 receptor alpha chain immunoreceptor tyrosine-based inhibitory motif - PubMed (original) (raw)
In vivo regulation of the allergic response by the IL-4 receptor alpha chain immunoreceptor tyrosine-based inhibitory motif
Raffi Tachdjian et al. J Allergy Clin Immunol. 2010 May.
Abstract
Background: Signaling by IL-4 and IL-13 through the IL-4 receptor alpha chain (IL-4Ralpha) plays a critical role in the pathology of allergic diseases. The IL-4Ralpha is endowed with an immunoreceptor tyrosine-based inhibitory motif (ITIM) centered on tyrosine 709 (Y709) in the cytoplasmic domain that binds a number of regulatory phosphatases. The function of the ITIM in the in vivo regulation of IL-4 receptor signaling remains unknown.
Objective: We sought to determine the in vivo function of the IL-4Ralpha ITIM by using mice in which the ITIM was inactivated by mutagenesis of the tyrosine Y709 residue into phenylalanine (F709).
Methods: F709 ITIM mutant mice were derived by means of knock-in mutagenesis. Activation of intracellular signaling cascades by IL-4 and IL-13 was assessed by means of intracellular staining of phosphorylated signaling intermediates and gene expression analysis. In vivo responses to allergic sensitization were assessed by using models of allergic airway inflammation.
Results: The F709 mutation increased signal transducer and activator of transcription 6 phosphorylation by IL-4 and, disproportionately, by IL-13. This was associated with exaggerated T(H)2 polarization, enhanced alternative macrophage activation by IL-13, augmented basal and antigen-induced IgE responses, and intensified allergen-induced eosinophilic airway inflammation and hyperreactivity.
Conclusions: These results point to a physiologic negative regulatory role for the Y709 ITIM in signaling through IL-4Ralpha, especially by IL-13.
Copyright 2010 American Academy of Allergy, Asthma & Immunology. Published by Mosby, Inc. All rights reserved.
Figures
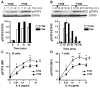
Fig. 1
Enhanced STAT6 activation in the C.129.Il4raF709/F709 mutant mice. A. Upper and middle panels: Western blot analysis of pSTAT6 induction in B cells at different time points following treatment with IL-4 (100 ng/ml). Lower panel: densitometric measurement of pSTAT6, normalized for STAT6 content. B. Upper panel: Decay of pSTAT6 signal in B cells upon cessation of IL-4 treatment. Cells were treated for 10 min with IL-4 (100 ng/ml), then washed (arrow) and analyzed at the indicated time points for pSTAT6 content. Lower panel: densitometric measurement of pSTAT, normalized for STAT6 content. C, D. Flow cytometric analysis of pSTAT6 induction in WT (Y709) and mutant (F709) B cells (C) or T cells (D) following treatment with graded concentrations of IL-4 for 15 min. Results for panels A and B are representative of 2 independent experiments each, and for C and D the averages of 3 mice per group/experiment with similar results observed in 3 independent experiments. ‡ p<0.0001 for genotype effect (F709 versus Y709) by repeat measure two way ANOVA; *p<0.05, **p<0.01 by two way ANOVA with Bonferroni post-test analysis of groups.
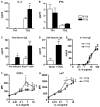
Fig. 2
The F709 mutation enhances IL-4 production by Th2 cells and IgE responses in vivo. A, B. Cytokine production by Th cells. Naive WT (Y709) and F709 splenic T cells (n=3-7 mice/group) were differentiated into Th1 (A) or Th2 (B) cells, then stimulated with anti-TCRβ mAB for 48 h. Culture supernatants were analyzed for IL-4 and IFNγ production by ELISA. C-D. Total and OVA-specific serum IgE levels in F709 and WT control mice (n=6-8/group) collected prior to (pre-immune) or following immunization with OVA/alum. E. In vitro IgE production by purified splenic B cells of WT and F709 mice (n=5/group) following stimulation with anti-CD40 mAb + IL-4. There was no significant difference between the two groups (repeat measure two way ANOVA). F, G. Upregulation of CD23 and I-Ad expression in Y709 and F709 B cells treated with increasing concentrations of IL-4 (n=3/group). *P<0.05, **P<0.01, ***P<0.001; †p<0.01, ‡p<0.0001 for genotype effect (Student's t-test for panel D, and 2 way ANOVA with post-test analysis for panels A, C, F and G).
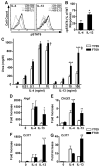
Fig. 3
The F709 mutation enhances alternative macrophage activation. A. Intracellular staining of pSTAT6 in WT (Y709) and F709 BMDM stimulated for 15 min at 37 °C with IL-4 (10 ng/ml) or IL-13 (100 ng/ml). B. Bar graph plot of the difference in the relative increase in pSTAT6 formation (ΔpSTAT6) in IL-4 and IL-13 treated F709 as compared to similarly treated Y709 BMDC (n=3 mice/group). C. Arginase activity in lysates of BMDMs treated with various concentrations of IL-4 or IL-13 and analyzed after 48 h by measurement of urea production ((n=9-12/group). D-G. Real-time PCR of genes encoding arginase 1 (Arg1), chitinase 3-like 3 (Chi3l3), and Ccl11 in BMDMs (D-F) and Ccl11 in primary lung fibroblast cultures (G) stimulated for 20 h with IL-4 or IL-13 at 10 ng/ml each (n=7-12/group). *p<0.05, **p<0.01, ***p<0.001; †p<0.01, ‡p<0.0001 for genotype effect (Student's t-test for panel B, and 2 way ANOVA with post-test analysis for panels C-G).
Fig 4
The F709 mutation impairs SHP-1 activation in BMDM cells. WT or F709 mutant BMDM cells were treated with IL-4 or IL-13 at 10 or 100 ng/ml and for 15 or 60 min, as indicated. Cellular lysates were derived and probed for pSHP-1, pSHP-2, pSHIP-1, pJAK1, pJAK3, and pTYK2, and β-Tubulin, the latter as a protein loading control. Results are representative of 2-3 independent experiments per immunblot panel.
Fig. 5
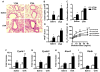
The Y709F substitution promotes acute antigen-induced allergic airway inflammation and AHR. Sham (Saline/Alum) or OVA/Alum sensitized C.129.Il4raY709/Y709 (Y709) and C.129.Il4raF709/F709 (F709) mice (n=10 mice/group) were challenged intranasally with OVA on 3 consecutive days then analyzed. A. Lung histology (PAS staining, 20×). B. Peribronchial inflammation scores. C. PAS-positive goblet cells in the airways. D. Eosinophilia in BAL fluid of sham and OVA immunized mice challenged with OVA. E. AHR measurements. Changes in lung resistance (RL) were measured in mice that had been either sham immunized or subjected to a limited protocol of OVA immunization (n=5-8/group). F-I. Real time PCR analysis of Cysltr1, Cysltr2, Alox5, Alox15, and Rtnlb mRNA levels in total lung extracts of OVA sensitized and challenged mice versus controls (n=3-6/group). *p<0.05, **p<0.01, ***p<0.001; ‡p<0.0001 for genotype effect (B: Kruskal Wallis test; C-I: 2 way ANOVA with post-test analysis).
Similar articles
- Immunoreceptor tyrosine-based inhibitory motif of the IL-4 receptor associates with SH2-containing phosphatases and regulates IL-4-induced proliferation.
Kashiwada M, Giallourakis CC, Pan PY, Rothman PB. Kashiwada M, et al. J Immunol. 2001 Dec 1;167(11):6382-7. doi: 10.4049/jimmunol.167.11.6382. J Immunol. 2001. PMID: 11714803 - IgE-mediated systemic anaphylaxis and impaired tolerance to food antigens in mice with enhanced IL-4 receptor signaling.
Mathias CB, Hobson SA, Garcia-Lloret M, Lawson G, Poddighe D, Freyschmidt EJ, Xing W, Gurish MF, Chatila TA, Oettgen HC. Mathias CB, et al. J Allergy Clin Immunol. 2011 Mar;127(3):795-805.e1-6. doi: 10.1016/j.jaci.2010.11.009. Epub 2010 Dec 17. J Allergy Clin Immunol. 2011. PMID: 21167580 Free PMC article. - Interleukin-4, interleukin-13, signal transducer and activator of transcription factor 6, and allergic asthma.
Kuperman DA, Schleimer RP. Kuperman DA, et al. Curr Mol Med. 2008 Aug;8(5):384-92. doi: 10.2174/156652408785161032. Curr Mol Med. 2008. PMID: 18691065 Free PMC article. Review. - Role of IgE in the development of allergic airway inflammation and airway hyperresponsiveness--a murine model.
Hamelmann E, Tadeda K, Oshiba A, Gelfand EW. Hamelmann E, et al. Allergy. 1999 Apr;54(4):297-305. doi: 10.1034/j.1398-9995.1999.00085.x. Allergy. 1999. PMID: 10371087 Review.
Cited by
- Regulatory T cells in dominant immunologic tolerance.
Georgiev P, Benamar M, Han S, Haigis MC, Sharpe AH, Chatila TA. Georgiev P, et al. J Allergy Clin Immunol. 2024 Jan;153(1):28-41. doi: 10.1016/j.jaci.2023.09.025. Epub 2023 Sep 29. J Allergy Clin Immunol. 2024. PMID: 37778472 Review. - Severe allergic dysregulation due to a gain of function mutation in the transcription factor STAT6.
Baris S, Benamar M, Chen Q, Catak MC, Martínez-Blanco M, Wang M, Fong J, Massaad MJ, Sefer AP, Kara A, Babayeva R, Eltan SB, Yucelten AD, Bozkurtlar E, Cinel L, Karakoc-Aydiner E, Zheng Y, Wu H, Ozen A, Schmitz-Abe K, Chatila TA. Baris S, et al. J Allergy Clin Immunol. 2023 Jul;152(1):182-194.e7. doi: 10.1016/j.jaci.2023.01.023. Epub 2023 Feb 8. J Allergy Clin Immunol. 2023. PMID: 36758835 Free PMC article. - Modulation of IL-4/IL-13 cytokine signaling in the context of allergic disease.
Shankar A, McAlees JW, Lewkowich IP. Shankar A, et al. J Allergy Clin Immunol. 2022 Aug;150(2):266-276. doi: 10.1016/j.jaci.2022.06.012. J Allergy Clin Immunol. 2022. PMID: 35934680 Free PMC article. Review. - IL-4 receptor alpha signaling alters oral food challenge and immunotherapy outcomes in mice.
Ganesan V, Sharma A, Tomar S, Schuler CF 4th, Hogan SP. Ganesan V, et al. J Allergy Clin Immunol. 2023 Jan;151(1):182-191.e6. doi: 10.1016/j.jaci.2022.07.011. Epub 2022 Aug 4. J Allergy Clin Immunol. 2023. PMID: 35934083 Free PMC article.
References
- Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–38. - PubMed
- Chatila TA. Interleukin-4 receptor signlaing pathways in asthma pathogenesis. Trends Mol Med. 2004;10:493–9. - PubMed
- Hershey GK. IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol. 2003;111:677–90. quiz 91. - PubMed
- Russell SM, Keegan AD, Harada N, Nakamura Y, Noguchi M, Leland P, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-4 receptor. Science. 1993;262:1880–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI054471/AI/NIAID NIH HHS/United States
- R01 AI065617-09S1/AI/NIAID NIH HHS/United States
- U19AI070453/AI/NIAID NIH HHS/United States
- R01 AI026322/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R21 AI080002/AI/NIAID NIH HHS/United States
- T32 HD007512/HD/NICHD NIH HHS/United States
- R01 AI065617-09/AI/NIAID NIH HHS/United States
- R01 AI065617-07/AI/NIAID NIH HHS/United States
- R21 AI080002-02/AI/NIAID NIH HHS/United States
- R01 AI065617/AI/NIAID NIH HHS/United States
- R21 AI087627-01/AI/NIAID NIH HHS/United States
- R01 AI065617-08/AI/NIAID NIH HHS/United States
- R01 AI065617-10/AI/NIAID NIH HHS/United States
- R21 AI080002-01/AI/NIAID NIH HHS/United States
- R21 AI087627/AI/NIAID NIH HHS/United States
- R01 AI065617-06A3/AI/NIAID NIH HHS/United States
- 2R01AI065617/AI/NIAID NIH HHS/United States
- R01AI054471/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials