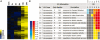agriGO: a GO analysis toolkit for the agricultural community - PubMed (original) (raw)
. 2010 Jul;38(Web Server issue):W64-70.
doi: 10.1093/nar/gkq310. Epub 2010 Apr 30.
Affiliations
- PMID: 20435677
- PMCID: PMC2896167
- DOI: 10.1093/nar/gkq310
agriGO: a GO analysis toolkit for the agricultural community
Zhou Du et al. Nucleic Acids Res. 2010 Jul.
Abstract
Gene Ontology (GO), the de facto standard in gene functionality description, is used widely in functional annotation and enrichment analysis. Here, we introduce agriGO, an integrated web-based GO analysis toolkit for the agricultural community, using the advantages of our previous GO enrichment tool (EasyGO), to meet analysis demands from new technologies and research objectives. EasyGO is valuable for its proficiency, and has proved useful in uncovering biological knowledge in massive data sets from high-throughput experiments. For agriGO, the system architecture and website interface were redesigned to improve performance and accessibility. The supported organisms and gene identifiers were substantially expanded (including 38 agricultural species composed of 274 data types). The requirement on user input is more flexible, in that user-defined reference and annotation are accepted. Moreover, a new analysis approach using Gene Set Enrichment Analysis strategy and customizable features is provided. Four tools, SEA (Singular enrichment analysis), PAGE (Parametric Analysis of Gene set Enrichment), BLAST4ID (Transfer IDs by BLAST) and SEACOMPARE (Cross comparison of SEA), are integrated as a toolkit to meet different demands. We also provide a cross-comparison service so that different data sets can be compared and explored in a visualized way. Lastly, agriGO functions as a GO data repository with search and download functions; agriGO is publicly accessible at http://bioinfo.cau.edu.cn/agriGO/.
Figures
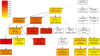
Figure 1.
Hierarchical tree graph of overrepresented GO terms in biological process category generated by SEA. Boxes in the graph represent GO terms labeled by their GO ID, term definition and statistical information. The significant term (adjusted P ≤ 0.05) are marked with color, while non-significant terms are shown as white boxes. The diagram, the degree of color saturation of a box is positively correlated to the enrichment level of the term. Solid, dashed, and dotted lines represent two, one and zero enriched terms at both ends connected by the line, respectively. The rank direction of the graph is set to from top to bottom.
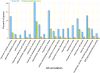
Figure 2.
Flash bar chart of overrepresented terms in all three categories. The _Y_-axis is the percentage of genes mapped by the term, and represents the abundance of the GO term. The percentage for the input list is calculated by the number of genes mapped to the GO term divided by the number of all genes in the input list. The same calculation was applied to the reference list to generate its percentage. These two lists are represented using different custom colors. The _X_-axis is the definition of GO terms.
Figure 3.
Hierarchical clustering of test-sample cluster and cross-comparison of its analysis results by PAGE in HTML table mode. (A) Experiments were performed with different cold treatment time (0.5, 1, 3, 6, 12 and 24 h) by AtGenExpress project (24). Probe set signal intensity was computed using RMA (33), and hierarchical clustering based on log2 Cold/CK ratio of the probe set at each time point was done by Cluster 3.0 (Cluster 3.0, command line version <
http://bonsai.ims.u-tokyo.ac.jp/mdehoon/software/cluster
>). For the test sample, 1921 probe sets showing coordinated upregulation at later time points of cold treatment were selected. (B) The 1921 probe sets in the test sample were analyzed by PAGE, and the comparison is displayed in HTML table mode. The colored blocks represent the level of up/downregulation of each term at a certain time-point. The yellow-to-red, cyan-to-blue and grayscale represent the term is upregulated, downregulated and non-significant, respectively. The adjusted _P-_value of the term determines the degree of color saturation of the corresponding box. Detailed information is provided for each term.
Similar articles
- agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update.
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z. Tian T, et al. Nucleic Acids Res. 2017 Jul 3;45(W1):W122-W129. doi: 10.1093/nar/gkx382. Nucleic Acids Res. 2017. PMID: 28472432 Free PMC article. - PlantGSEA: a gene set enrichment analysis toolkit for plant community.
Yi X, Du Z, Su Z. Yi X, et al. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W98-103. doi: 10.1093/nar/gkt281. Epub 2013 Apr 30. Nucleic Acids Res. 2013. PMID: 23632162 Free PMC article. - GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis.
Zheng Q, Wang XJ. Zheng Q, et al. Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W358-63. doi: 10.1093/nar/gkn276. Epub 2008 May 16. Nucleic Acids Res. 2008. PMID: 18487275 Free PMC article. - EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species.
Zhou X, Su Z. Zhou X, et al. BMC Genomics. 2007 Jul 24;8:246. doi: 10.1186/1471-2164-8-246. BMC Genomics. 2007. PMID: 17645808 Free PMC article. - Gene Ontology annotation of the rice blast fungus, Magnaporthe oryzae.
Meng S, Brown DE, Ebbole DJ, Torto-Alalibo T, Oh YY, Deng J, Mitchell TK, Dean RA. Meng S, et al. BMC Microbiol. 2009 Feb 19;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2180-9-S1-S8. BMC Microbiol. 2009. PMID: 19278556 Free PMC article. Review.
Cited by
- Genome-wide identification and characterization of microRNA genes and their targets in flax (Linum usitatissimum): Characterization of flax miRNA genes.
Barvkar VT, Pardeshi VC, Kale SM, Qiu S, Rollins M, Datla R, Gupta VS, Kadoo NY. Barvkar VT, et al. Planta. 2013 Apr;237(4):1149-61. doi: 10.1007/s00425-012-1833-5. Epub 2013 Jan 6. Planta. 2013. PMID: 23291876 - Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis.
Lu P, Han X, Qi J, Yang J, Wijeratne AJ, Li T, Ma H. Lu P, et al. Genome Res. 2012 Mar;22(3):508-18. doi: 10.1101/gr.127522.111. Epub 2011 Nov 21. Genome Res. 2012. PMID: 22106370 Free PMC article. - Experimental sink removal induces stress responses, including shifts in amino acid and phenylpropanoid metabolism, in soybean leaves.
Turner GW, Cuthbertson DJ, Voo SS, Settles ML, Grimes HD, Lange BM. Turner GW, et al. Planta. 2012 May;235(5):939-54. doi: 10.1007/s00425-011-1551-4. Epub 2011 Nov 23. Planta. 2012. PMID: 22109846 Free PMC article. - Miniature Seed6, encoding an endoplasmic reticulum signal peptidase, is critical in seed development.
Yi F, Gu W, Li J, Chen J, Hu L, Cui Y, Zhao H, Guo Y, Lai J, Song W. Yi F, et al. Plant Physiol. 2021 Apr 2;185(3):985-1001. doi: 10.1093/plphys/kiaa060. Plant Physiol. 2021. PMID: 33793873 Free PMC article. - Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum).
Wang J, Hu Z, Zhao T, Yang Y, Chen T, Yang M, Yu W, Zhang B. Wang J, et al. BMC Genomics. 2015 Feb 5;16(1):39. doi: 10.1186/s12864-015-1249-2. BMC Genomics. 2015. PMID: 25652024 Free PMC article.
References
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. - PubMed
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. - PubMed
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials