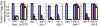Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity - PubMed (original) (raw)
Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity
Sivan Henis-Korenblit et al. Proc Natl Acad Sci U S A. 2010.
Abstract
When unfolded proteins accumulate in the endoplasmic reticulum (ER), the unfolded protein response is activated. This ER stress response restores ER homeostasis by coordinating processes that decrease translation, degrade misfolded proteins, and increase the levels of ER-resident chaperones. Ribonuclease inositol-requiring protein-1 (IRE-1), an endoribonuclease that mediates unconventional splicing, and its target, the XBP-1 transcription factor, are key mediators of the unfolded protein response. In this study, we show that in Caenorhabditis elegans insulin/IGF-1 pathway mutants, IRE-1 and XBP-1 promote lifespan extension and enhance resistance to ER stress. We show that these effects are not achieved simply by increasing the level of spliced xbp-1 mRNA and expression of XBP-1's normal target genes. Instead, in insulin/IGF-1 pathway mutants, XBP-1 collaborates with DAF-16, a FOXO-transcription factor that is activated in these mutants, to enhance ER stress resistance and to activate new genes that promote longevity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Fig. 1.
ire-1 and xbp-1 contribute to the longevity of daf-2(−) animals. (A) Reduction of ire-1 activity by the null mutation ok799 shortened the lifespan of daf-2(e1370) mutants more than it shortened the lifespan of wild type. (B) The xbp-1 (zc12) mutation shortened the lifespan of daf-2(e1370) mutants more than it shortened wild-type lifespan. (C) The xbp-1(zc12) mutation did not further shorten the lifespan of ire-1(−); daf-2(−) double mutants, suggesting that these genes function in a common pathway. [Compare survival curves of ire-1(−); daf-2(−) and ire-1(−); daf-2(−) xbp-1(zc12) mutants.] Note that xbp-1(zc12) mutation shortened the lifespan of daf-2(e1370) mutants, but not as much as did an ire-1(ok799) deletion. [Compare survival curves of ire-1(−); daf-2(−) and daf-2(−) xbp-1(zc12) mutants.] As xbp-1(zc12) is likely to eliminate xbp-1 function, this finding implies that ire-1 contributes to the longevity of daf-2 mutants in part through an _xbp-1_–independent pathway. For additional lifespan data, see
Table S1
. Also, see
Fig. S1
for data showing that other ER stress–response genes, namely abu-11, pek-1, and atf-6, are not involved in this pathway.
Fig. 2.
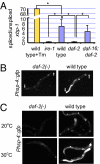
The ire-1/xbp-1 pathway is set at a lower level in daf-2(−) mutants. (A) Bar graph representing the mean ratio of spliced/unspliced xbp-1 transcripts, multiplied by a factor of 100, in three independent biological experiments. Control lane (yellow bar) presents the xbp-1 transcripts ratio in wild-type animals treated with tunicamycin. The rest of the bars present steady-state xbp-1 transcripts ratio in day-1 wild-type, ire-1, daf-2, and daf-16; daf-2 animals. These animals have not been treated with tunicamycin (blue bars). Error bars represent SEM. Asterisks mark Student t test values of P < 0.01. See
Fig. S2
for data showing a representative RT-PCR experiment. (B) Representative fluorescence micrographs (100-fold magnification) of day-3 adults harboring an integrated Phsp-4::gfp transgene in a wild-type or daf-2(e1370) background. daf-2 mutants expressed lower levels of the Phsp-4::gfp transgene than did wild type. wild type, n = 9, relative mean fluorescence intensity (m) = 1 ± 0.14; daf-2(e1370), n = 10, m = 0.34 ± 0.07, P = 0.0012. (C) daf-2(e1370) mutants expressed less Phsp-4::gfp transgene than did wild type even upon heat shock. Phsp-4::gfp expression at 20 °C or upon 3 h of heat shock at 30 °C. Arrows point at anterior and posterior ends of the animals.
Fig. 3.
daf-2(−) mutants are resistant to ER stress. Eggs from wild-type or daf-2(e1370) animals containing xbp-1(zc12), ire-1(ok799), pek-1(ok275), or atf-6(ok551) mutations were grown in the presence of 0, 2, or 5 μg/mL tunicamycin (corresponding to the blue, red, and off-white bars in the graph). Percentage of eggs that developed into mature adults was scored. Student t test values were calculated between daf-2(+) and daf-2(−) backgrounds. Each experiment was repeated independently with similar effects. Each strain was scored on three independent plates. Error bars represent SEM for repeat plates within the experiment. Asterisks mark Student t test values of P < 0.01.
Fig. 4.
dox-1 promotes longevity, but not ER stress resistance, in daf-2 mutants. (A) Reduction of dox-1 activity by RNAi shortened the lifespan of daf-2(mu150) mutants more than it shortened the lifespan of daf-2(+) animals. Animals were in a fer-15(b26); fem-1(hc17) background to prevent reproduction. Similar results were seen in daf-2(e1370) mutants (
Table S1
). (B) Reduction of dox-1 did not increase the set-point of the ire-1/xbp-1 branch of the UPR in daf-2 mutants. Bar graph showing the mean intensity of day-2 adults expressing a Phsp-4::gfp transgene. daf-2(e1370), n = 13, relative mean fluorescence intensity (m) = 21.9 ± 2.2; dox-1(RNAi); daf-2(e1370), n = 17, m = 23.3 ± 2.3, P = 0.677. See
Fig. S4
for representative micrographs and wild-type data. (C) dox-1 expression was up-regulated in daf-2 mutants. Bar graph showing the fraction of animals expressing high levels of Pdox-1::gfp. Day-1 adults of each strain were classified as expressing high or low GFP levels (
Fig. S4
). The fraction of animals expressing high levels of Pdox-1::gfp was 1.35 fold higher in transgenic daf-2(e1370) mutants than in wild-type animals (P = 0.037), but was not significantly increased in daf-2(e1370) xbp-1(tm2457) double mutants. The fraction of animals expressing high levels of Pdox-1::gfp was 1.52 fold higher in transgenic daf-2(e1368) mutants than in wild-type animals (P = 0.007), but was not increased in daf-2(e1368) xbp-1(tm2457) double mutants. The fraction of animals expressing high levels of Pdox-1::gfp was higher by 1.8 fold in transgenic daf-2(e1370) mutants than in wild-type animals (P = 0.026), but was not significantly increased in daf-16 (mu86); daf-2(e1370) double mutants. Error bars represent SEM of at least two independent trials. Asterisks mark Student t test values of P < 0.05.
Fig. 5.
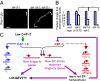
daf-16 contributes to the ER stress resistance in daf-2 mutants. (A) Representative fluorescence micrographs of day-1 adults harboring an integrated Phsp-4::gfp transgene in a daf-2(e1370) or daf-16(mu86); daf-2(e1370) background. daf-16 deletion (and inactivation of daf-16 by RNAi) increased the level of Phsp-4::gfp in daf-2(−) mutants. daf-2(e1370), n = 20, relative mean fluorescence intensity (m) = 1 ± 0.06; daf-16(mu86); daf-2(e1370), n = 28, m = 2.70 ± 0.2, P < 0.0001. Error bars represent SEM. Arrows point at anterior and posterior ends of the animals. (B) Tunicamycin resistance developmental assay. Eggs from wild-type, daf-2(e1370), and daf-16(mu86); daf-2(e1370) animals were grown in the presence of 0 or 5 μg/mL tunicamycin (corresponding to the blue and off-white bars in the graph). Percentage of eggs that developed into mature adults was scored. Student t test values were calculated relative to daf-16(−); daf-2(−). Asterisks mark Student t test values of P < 0.05. This experiment was repeated independently with similar effects. (C) Model for how XBP-1 extends lifespan, increases ER stress resistance, and also lowers XBP-1 levels when insulin/IGF-1 signaling is reduced. In daf-2 mutants, multiple transcription factors, such as DAF-16/FOXO, HSF-1, ELT-3, and SKN-1 are activated. These can act in a combinatorial fashion with XBP-1 to turn on new longevity genes and new ER stress response genes (both in purple), which, unlike the “normal” ER stress–protective genes (in red), cannot be induced by XBP-1 alone. This combinatorial control results in a wider array of longevity and ER stress–protective genes, both of which contribute to the longevity of daf-2 mutants. In addition, as part of a negative feedback loop, the wider array of ER stress protective genes expressed in daf-2 mutants by xbp-1 can better reduce the level of unfolded proteins in the ER and improve ER homeostasis. Improved ER homeostasis would, in turn, dictate a lower basal activity level of the ire-1/xbp-1 pathway. In this model, the enhanced ER stress resistance of daf-2 mutants is completely _xbp-1_–dependent, and yet, because the ER stress response is more efficient, the set point of XBP-1 and its normal downstream targets is lower than in wild type.
Similar articles
- Reduced Insulin/Insulin-Like Growth Factor Receptor Signaling Mitigates Defective Dendrite Morphogenesis in Mutants of the ER Stress Sensor IRE-1.
Salzberg Y, Coleman AJ, Celestrin K, Cohen-Berkman M, Biederer T, Henis-Korenblit S, Bülow HE. Salzberg Y, et al. PLoS Genet. 2017 Jan 23;13(1):e1006579. doi: 10.1371/journal.pgen.1006579. eCollection 2017 Jan. PLoS Genet. 2017. PMID: 28114319 Free PMC article. - Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism.
Stout GJ, Stigter EC, Essers PB, Mulder KW, Kolkman A, Snijders DS, van den Broek NJ, Betist MC, Korswagen HC, Macinnes AW, Brenkman AB. Stout GJ, et al. Mol Syst Biol. 2013 Jul 2;9:679. doi: 10.1038/msb.2013.35. Mol Syst Biol. 2013. PMID: 23820781 Free PMC article. - PEK-1 is crucial for hormesis induced by inhibition of the IRE-1/XBP-1 pathway in the Caenorhabditis elegans mev-1 mutant.
Eisermann DJ, Wenzel U, Fitzenberger E. Eisermann DJ, et al. Biochem Biophys Res Commun. 2016 May 13;473(4):1052-1057. doi: 10.1016/j.bbrc.2016.04.014. Epub 2016 Apr 5. Biochem Biophys Res Commun. 2016. PMID: 27055592 - Vitamin D Promotes Protein Homeostasis and Longevity via the Stress Response Pathway Genes skn-1, ire-1, and xbp-1.
Mark KA, Dumas KJ, Bhaumik D, Schilling B, Davis S, Oron TR, Sorensen DJ, Lucanic M, Brem RB, Melov S, Ramanathan A, Gibson BW, Lithgow GJ. Mark KA, et al. Cell Rep. 2016 Oct 25;17(5):1227-1237. doi: 10.1016/j.celrep.2016.09.086. Cell Rep. 2016. PMID: 27783938 Free PMC article. - The search for DAF-16/FOXO transcriptional targets: approaches and discoveries.
Murphy CT. Murphy CT. Exp Gerontol. 2006 Oct;41(10):910-21. doi: 10.1016/j.exger.2006.06.040. Epub 2006 Aug 24. Exp Gerontol. 2006. PMID: 16934425 Review.
Cited by
- Inactivation of the Caenorhabditis elegans RNF-5 E3 ligase promotes IRE-1-independent ER functions.
Adir O, Bening-Abu-Shach U, Arbib S, Henis-Korenblit S, Broday L. Adir O, et al. Autophagy. 2021 Sep;17(9):2401-2414. doi: 10.1080/15548627.2020.1827778. Epub 2020 Oct 15. Autophagy. 2021. PMID: 32981418 Free PMC article. - Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways.
Dues DJ, Andrews EK, Schaar CE, Bergsma AL, Senchuk MM, Van Raamsdonk JM. Dues DJ, et al. Aging (Albany NY). 2016 Apr;8(4):777-95. doi: 10.18632/aging.100939. Aging (Albany NY). 2016. PMID: 27053445 Free PMC article. - Insulin-like growth factor 2 (IGF2) protects against Huntington's disease through the extracellular disposal of protein aggregates.
García-Huerta P, Troncoso-Escudero P, Wu D, Thiruvalluvan A, Cisternas-Olmedo M, Henríquez DR, Plate L, Chana-Cuevas P, Saquel C, Thielen P, Longo KA, Geddes BJ, Lederkremer GZ, Sharma N, Shenkman M, Naphade S, Sardi SP, Spichiger C, Richter HG, Court FA, Tshilenge KT, Ellerby LM, Wiseman RL, Gonzalez-Billault C, Bergink S, Vidal RL, Hetz C. García-Huerta P, et al. Acta Neuropathol. 2020 Nov;140(5):737-764. doi: 10.1007/s00401-020-02183-1. Epub 2020 Jul 8. Acta Neuropathol. 2020. PMID: 32642868 Free PMC article. - Application of Caenorhabditis elegans for Research on Endoplasmic Reticulum Stress.
Xu Y, Park Y. Xu Y, et al. Prev Nutr Food Sci. 2018 Dec;23(4):275-281. doi: 10.3746/pnf.2018.23.4.275. Epub 2018 Dec 31. Prev Nutr Food Sci. 2018. PMID: 30675455 Free PMC article. Review. - Proteostasis and longevity: when does aging really begin?
Labbadia J, Morimoto RI. Labbadia J, et al. F1000Prime Rep. 2014 Feb 3;6:7. doi: 10.12703/P6-7. eCollection 2014. F1000Prime Rep. 2014. PMID: 24592319 Free PMC article. Review.
References
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. - PubMed
- Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. - PubMed
- Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous