eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation - PubMed (original) (raw)
eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation
Martin D Jennings et al. Nature. 2010.
Erratum in
- Nature. 2010 Nov 4;468(7320):122
Abstract
In protein synthesis initiation, the eukaryotic translation initiation factor (eIF) 2 (a G protein) functions in its GTP-bound state to deliver initiator methionyl-tRNA (tRNA(i)(Met)) to the small ribosomal subunit and is necessary for protein synthesis in all cells. Phosphorylation of eIF2 [eIF2(alphaP)] is critical for translational control in diverse settings including nutrient deprivation, viral infection and memory formation. eIF5 functions in start site selection as a GTPase accelerating protein (GAP) for the eIF2.GTP.tRNA(i)(Met) ternary complex within the ribosome-bound pre-initiation complex. Here we define new regulatory functions of eIF5 in the recycling of eIF2 from its inactive eIF2.GDP state between successive rounds of translation initiation. First we show that eIF5 stabilizes the binding of GDP to eIF2 and is therefore a bi-functional protein that acts as a GDP dissociation inhibitor (GDI). We find that this activity is independent of the GAP function and identify conserved residues within eIF5 that are necessary for this role. Second we show that eIF5 is a critical component of the eIF2(alphaP) regulatory complex that inhibits the activity of the guanine-nucleotide exchange factor (GEF) eIF2B. Together our studies define a new step in the translation initiation pathway, one that is critical for normal translational controls.
Figures
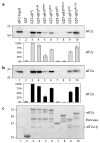
Figure 1. eIF5 has GDI activity
a) Scheme for GDI activity assay. b) Increasing eIF5 stabilises GDP-binding to eIF2. Koff GDP from 60 pmol eIF2 with varying concentrations of GST-eIF5 (0-240 pmol, open circles) or GST alone (filled circle). Molar eIF2:GST-eIF5 protein ratios are indicated. c) Defining regions required for GDI activity. Mean Koff GDP (60 pmol eIF2) for indicated constructs derived from reactions with GST- or FLAG-eIF5 proteins (120 pmol). Black bars represent a significant reduction in Koff GDP (P<0.0001, unpaired Student's _t_-test). Errors show standard deviation (n>3). 2.9 mM Mg2+ was used in b) and c).
Figure 2. The CTD of eIF5 is critical for interaction with eIF2
Affinity chromatography assay between eIF2 (110 pmol) and the indicated immobilized GST-eIF5 constructs. eIF2 was detected by immunoblotting using antibodies specific for a) eIF2γ or b) eIF2α. Representative blots are shown. Signal intensity was quantified (Adobe Photoshop) and the mean ± standard deviation (n=3) are shown below. c) Total protein in each sample stained with Ponceau S. Inputs (lanes 1) represent 10% of total.
Figure 3. The Linker Region of eIF5 interacts with γ subunit of eIF2
Affinity chromatography as in figure 2 between indicated immobilized GST-eIF5 constructs and total cell extracts (500 μg) expressing c-Myc-6xHis-eIF2γ from either a single copy (sc) or high copy (hc) plasmid. Immunoblots were developed with c-Myc and eIF2α antibodies. Total protein in each sample was stained with Ponceau S. Inputs (lane 1) represent 5% (blots) or 1% (stain) of total.
Figure 4. GDI activity antagonises eIF2B and affects GCN4 activation in vivo
a-c) Strains expressing single (sc) or high copy (hc) eIF5 plasmids as the source of eIF5 and co-transformed with plasmids expressing GCN2, vector alone (_gcn2_Δ) [a, b] or the constitutively active mutant GCN2M788V,E1591K (GCN2c) [c] were grown as stated. d) Left, immunoblots following FLAG-eIF5 immune precipitation of protein complexes from cells grown in nutrient sufficient conditions (SCD) and following starvation (SD+3AT). Right, quantification ± standard deviation (n=3). e) Model for recycling and regulation of eIF2, incorporating eIF5 GDI activity. Dashed grey arrows indicate steps that limit eIF2 recycling.
Similar articles
- eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation.
Jennings MD, Zhou Y, Mohammad-Qureshi SS, Bennett D, Pavitt GD. Jennings MD, et al. Genes Dev. 2013 Dec 15;27(24):2696-707. doi: 10.1101/gad.231514.113. Genes Dev. 2013. PMID: 24352424 Free PMC article. - Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2.
Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Asano K, et al. EMBO J. 1999 Mar 15;18(6):1673-88. doi: 10.1093/emboj/18.6.1673. EMBO J. 1999. PMID: 10075937 Free PMC article. - eIF2β is critical for eIF5-mediated GDP-dissociation inhibitor activity and translational control.
Jennings MD, Kershaw CJ, White C, Hoyle D, Richardson JP, Costello JL, Donaldson IJ, Zhou Y, Pavitt GD. Jennings MD, et al. Nucleic Acids Res. 2016 Nov 16;44(20):9698-9709. doi: 10.1093/nar/gkw657. Epub 2016 Jul 25. Nucleic Acids Res. 2016. PMID: 27458202 Free PMC article. - Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation.
Das S, Maitra U. Das S, et al. Prog Nucleic Acid Res Mol Biol. 2001;70:207-31. doi: 10.1016/s0079-6603(01)70018-9. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11642363 Review. - A new function and complexity for protein translation initiation factor eIF2B.
Jennings MD, Pavitt GD. Jennings MD, et al. Cell Cycle. 2014;13(17):2660-5. doi: 10.4161/15384101.2014.948797. Cell Cycle. 2014. PMID: 25486352 Free PMC article. Review.
Cited by
- Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5.
Loughran G, Sachs MS, Atkins JF, Ivanov IP. Loughran G, et al. Nucleic Acids Res. 2012 Apr;40(7):2898-906. doi: 10.1093/nar/gkr1192. Epub 2011 Dec 7. Nucleic Acids Res. 2012. PMID: 22156057 Free PMC article. - The metaphorical swiss army knife: The multitude and diverse roles of HEAT domains in eukaryotic translation initiation.
Friedrich D, Marintchev A, Arthanari H. Friedrich D, et al. Nucleic Acids Res. 2022 Jun 10;50(10):5424-5442. doi: 10.1093/nar/gkac342. Nucleic Acids Res. 2022. PMID: 35552740 Free PMC article. Review. - Regulation and function of elF2B in neurological and metabolic disorders.
Hanson FM, Hodgson RE, de Oliveira MIR, Allen KE, Campbell SG. Hanson FM, et al. Biosci Rep. 2022 Jun 30;42(6):BSR20211699. doi: 10.1042/BSR20211699. Biosci Rep. 2022. PMID: 35579296 Free PMC article. Review. - Structural basis for the inhibition of translation through eIF2α phosphorylation.
Gordiyenko Y, Llácer JL, Ramakrishnan V. Gordiyenko Y, et al. Nat Commun. 2019 Jun 14;10(1):2640. doi: 10.1038/s41467-019-10606-1. Nat Commun. 2019. PMID: 31201334 Free PMC article. - In vivo transcriptional analysis of mice infected with Leishmania major unveils cellular heterogeneity and altered transcriptomic profiling at single-cell resolution.
Venugopal G, Bird JT, Washam CL, Roys H, Bowlin A, Byrum SD, Weinkopff T. Venugopal G, et al. PLoS Negl Trop Dis. 2022 Jul 5;16(7):e0010518. doi: 10.1371/journal.pntd.0010518. eCollection 2022 Jul. PLoS Negl Trop Dis. 2022. PMID: 35789215 Free PMC article.
References
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. - PubMed
- Scheuner D, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 2005;11:757–764. - PubMed
- Mohr I. Neutralizing innate host defenses to control viral translation in HSV-1 infected cells. Int. Rev. Immunol. 2004;23:199–220. - PubMed
Additional Reference
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBE0020051/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E002005/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H010599/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous